Navigation auf uzh.ch
Navigation auf uzh.ch
Muskalla L, Güldenpfennig A, Hottiger MO
Subcellular Quantitation of ADP-Ribosylation by High-Content Microscopy
Methods Mol Biol. 2023;2609:101-109. doi: 10.1007/978-1-0716-2891-1_7. PMID: 36515832.
Schraml P, Aimi F, Zoche M, Aguilera-Garcia D, Arnold F, Moch H, Hottiger MO
Altered cytoplasmic and nuclear ADP-ribosylation levels analyzed with an improved ADP-ribose binder are a prognostic factor in renal cell carcinoma
J Pathol Clin Res. 2023 Mar 31. doi: 10.1002/cjp2.320. Epub ahead of print. PMID: 36999983.
Lüscher B, Ahel I, Altmeyer M, Ashworth A, Bai P, Chang P, Cohen M, Corda D, Dantzer F, Daugherty MD, Dawson TM, Dawson VL, Deindl S, Fehr AR, Feijs KLH, Filippov DV, Gagné JP, Grimaldi G, Guettler S, Hoch NC, Hottiger MO, Korn P, Kraus WL, Ladurner A, Lehtiö L, Leung AKL, Lord CJ, Mangerich A, Matic I, Matthews J, Moldovan GL, Moss J, Natoli G, Nielsen ML, Niepel M, Nolte F, Pascal J, Paschal BM, Pawłowski K, Poirier GG, Smith S, Timinszky G, Wang ZQ, Yélamos J, Yu X, Zaja R, Ziegler M
ADP-ribosyltransferases, an update on function and nomenclature
Liu L, Sandow JJ, Leslie Pedrioli DM, Samson AL, Silke N, Kratina T, Ambrose RL, Doerflinger M, Hu Z, Morrish E, Chau D, Kueh AJ, Fitzibbon C, Pellegrini M, Pearson JS, Hottiger MO, Webb AI, Lalaoui N, Silke J
Tankyrase-mediated ADP-ribosylation is a regulator of TNF-induced death
Kuraoka S, Higashi H, Yanagihara Y, Sonawane AR, Mukai S, Mlynarchik AK, Whelan MC, Hottiger MO, Nasir W, Delanghe B, Aikawa M, Singh SA
A Novel Spectral Annotation Strategy Streamlines Reporting of Mono-ADP-ribosylated Peptides Derived from Mouse Liver and Spleen in Response to IFN-γ
Boehi F, Manetsch P, Hottiger MO
Interplay between ADP-ribosyltransferases and essential cell signaling pathways controls cellular responses
Aimi F, Moch H, Schraml P, Hottiger MO
Cytoplasmic ADP-ribosylation levels correlate with markers of patient outcome in distinct human cancers
Leutert M, Duan Y, Winzer R, Menzel S, Tolosa E, Magnus T, Hottiger MO, Koch-Nolte F, Rissiek B
Identification of the Mouse T Cell ADP-Ribosylome Uncovers ARTC2.2 Mediated Regulation of CD73 by ADP-Ribosylation
Hopp AK, Hottiger MO
Investigation of Mitochondrial ADP-Ribosylation Via Immunofluorescence
Methods Mol Biol. 2021;2276:165-171. doi: 10.1007/978-1-0716-1266-8_12. PMID: 34060040.
Lüthi SC, Howald A, Nowak K, Graage R, Bartolomei G, Neupert C, Sidler X, Leslie Pedrioli D, Hottiger MO
Establishment of a Mass-Spectrometry-Based Method for the Identification of the In Vivo Whole Blood and Plasma ADP-Ribosylomes
Bisceglie L, Hopp AK, Gunasekera K, Wright RH, Le Dily F, Vidal E, Dall'Agnese A, Caputo L, Nicoletti C, Puri PL, Beato M, Hottiger MO
MyoD induces ARTD1 and nucleoplasmic poly-ADP-ribosylation during fibroblast to myoblast transdifferentiation
Hopp AK, Hottiger MO
Uncovering the Invisible: Mono-ADP-ribosylation Moved into the Spotlight
Cells. 2021 Mar 19;10(3):680. doi: 10.3390/cells10030680. PMID: 33808662; PMCID: PMC8003356.
Hopp AK, Teloni F, Bisceglie L, Gondrand C, Raith F, Nowak K, Muskalla L, Howald A, Pedrioli PGA, Johnsson K, Altmeyer M, Pedrioli DML, Hottiger MO
Mitochondrial NAD+ Controls Nuclear ARTD1-Induced ADP-Ribosylation
Gehrig PM, Nowak K, Panse C, Leutert M, Grossmann J, Schlapbach R, Hottiger MO
Gas-Phase Fragmentation of ADP-Ribosylated Peptides: Arginine-Specific Side-Chain Losses and Their Implication in Database Searches
Glaus Garzon JF, Pastrello C, Jurisica I, Hottiger MO, Wenger RH, Borsig L
Tumor cell endogenous HIF-1α activity induces aberrant angiogenesis and interacts with TRAF6 pathway required for colorectal cancer development
Cheng D, Gao G, Di Lorenzo A, Jayne S, Hottiger MO, Richard S, Bedford MT
Genetic evidence for partial redundancy between the arginine methyltransferases CARM1 and PRMT6
Niu Y, Yang L, Gao T, Dong C, Zhang B, Yin P, Hopp AK, Li D, Gan R, Wang H, Liu X, Cao X, Xie Y, Meng X, Deng H, Zhang X, Ren J, Hottiger MO, Chen Z, Zhang Y, Liu X, Feng Y
A Type I-F Anti-CRISPR Protein Inhibits the CRISPR-Cas Surveillance Complex by ADP-Ribosylation
Nowak K, Rosenthal F, Karlberg T, Bütepage M, Thorsell AG, Dreier B, Grossmann J, Sobek J, Imhof R, Lüscher B, Schüler H, Plückthun A, Leslie Pedrioli DM, Hottiger MO
Engineering Af1521 improves ADP-ribose binding and identification of ADP-ribosylated proteins
Vdovenko D, Bachmann M, Wijnen WJ, Hottiger MO, Eriksson U, Valaperti A
The adaptor protein c-Cbl-associated protein (CAP) limits pro-inflammatory cytokine expression by inhibiting the NF-κB pathway
Ashok Y, Miettinen M, Oliveira DKH, Tamirat MZ, Näreoja K, Tiwari A, Hottiger MO, Johnson MS, Lehtiö L, Pulliainen AT
Discovery of Compounds Inhibiting the ADP-Ribosyltransferase Activity of Pertussis Toxin
Wang C, Xiao J, Nowak K, Gunasekera K, Alippe Y, Speckman S, Yang T, Kress D, Abu-Amer Y, Hottiger MO, Mbalaviele G
PARP1 Hinders Histone H2B Occupancy at the NFATc1 Promoter to Restrain Osteoclast Differentiation
Arsiwala T, Pahla J, van Tits LJ, Bisceglie L, Gaul DS, Costantino S, Miranda MX, Nussbaum K, Stivala S, Blyszczuk P, Weber J, Tailleux A, Stein S, Paneni F, Beer JH, Greter M, Becher B, Mostoslavsky R, Eriksson U, Staels B, Auwerx J, Hottiger MO, Lüscher TF, Matter CM
Sirt6 deletion in bone marrow-derived cells increases atherosclerosis - Central role of macrophage scavenger receptor 1
Hopp AK, Grüter P, Hottiger MO
Regulation of Glucose Metabolism by NAD+ and ADP-Ribosylation
Xu Y, Zhou P, Cheng S, Lu Q, Nowak K, Hopp AK, Li L, Shi X, Zhou Z, Gao W, Li D, He H, Liu X, Ding J, Hottiger MO, Shao F
A Bacterial Effector Reveals the V-ATPase-ATG16L1 Axis that Initiates Xenophagy
Cell. 2019 Jul 25;178(3):552-566.e20. doi: 10.1016/j.cell.2019.06.007
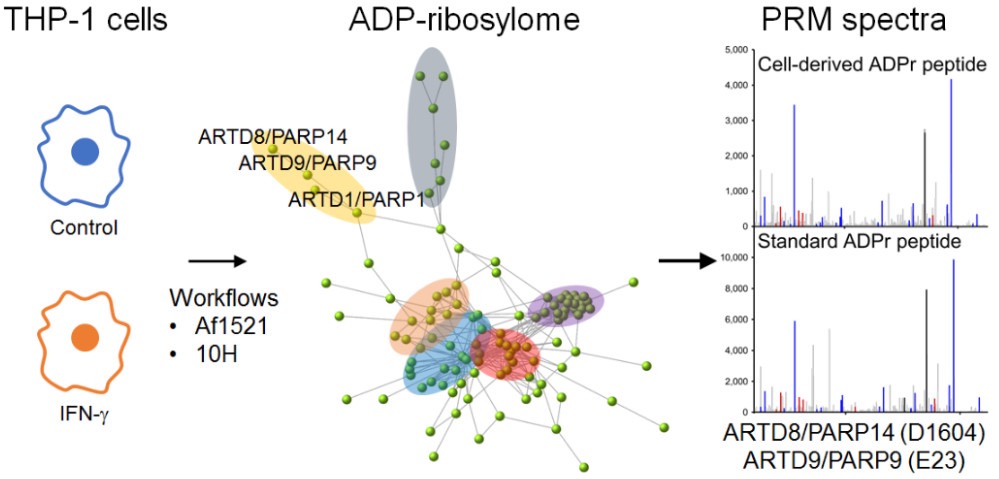
Higashi H, Maejima T, Lee LH, Yamazaki Y, Hottiger MO, Singh SA, Aikawa M
A Study into the ADP-Ribosylome of IFN-γ-Stimulated THP-1 Human Macrophage-like Cells Identifies ARTD8/PARP14 and ARTD9/PARP9 ADP-Ribosylation
J Proteome Res. 2019 Mar 21. doi:0.1021/acs.jproteome.8b00895
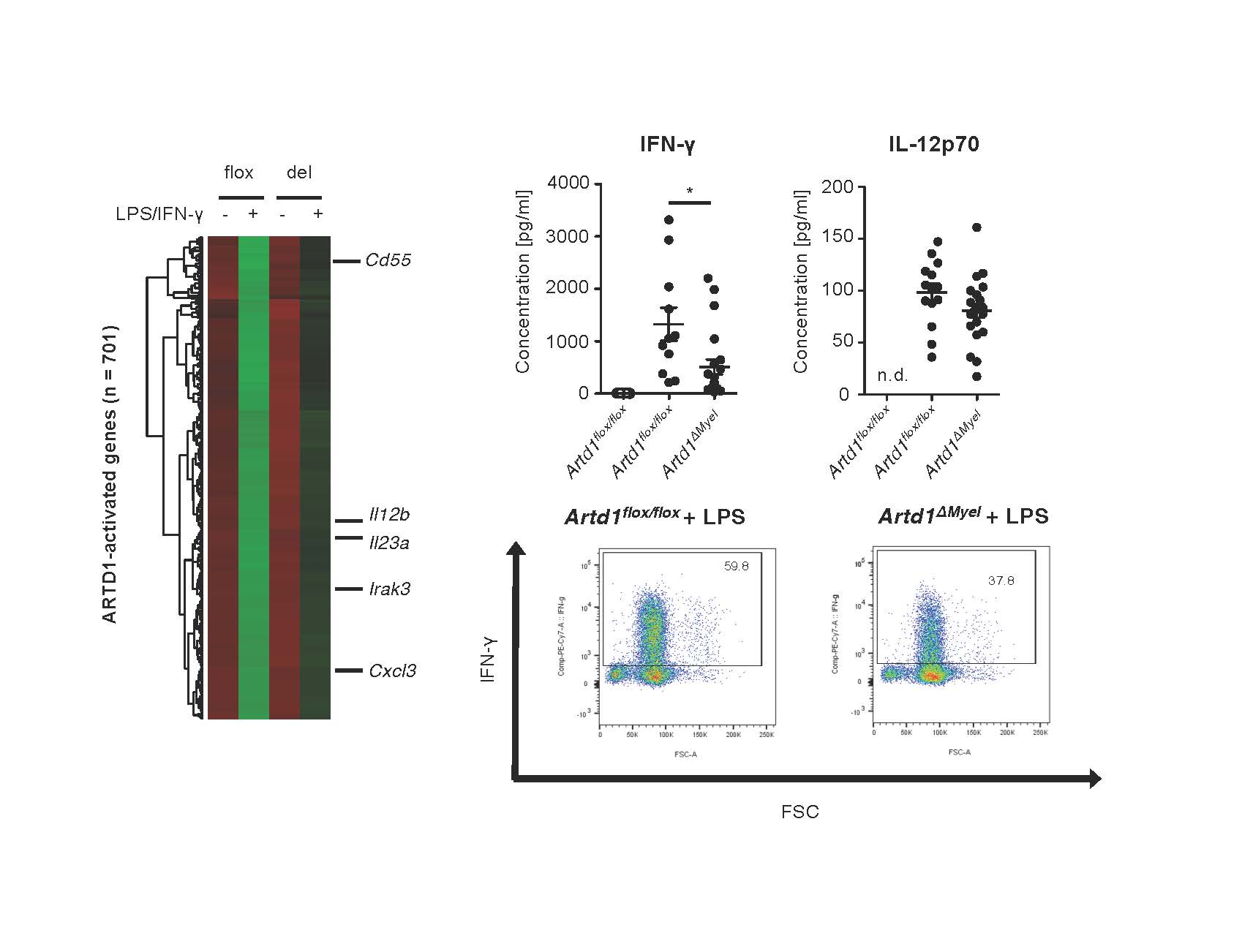
Kunze FA, Bauer M, Komuczki J, Lanzinger M, Gunasekera K, Hopp AK, Lehmann M, Becher B, Müller A, Hottiger MO
ARTD1 in Myeloid Cells Controls the IL-12/18-IFN-γ Axis in a Model of Sterile Sepsis, Chronic Bacterial Infection, and Cancer
J Immunol. 2019 Jan 23. pii: ji1801107. doi: 10.4049/jimmunol.1801107
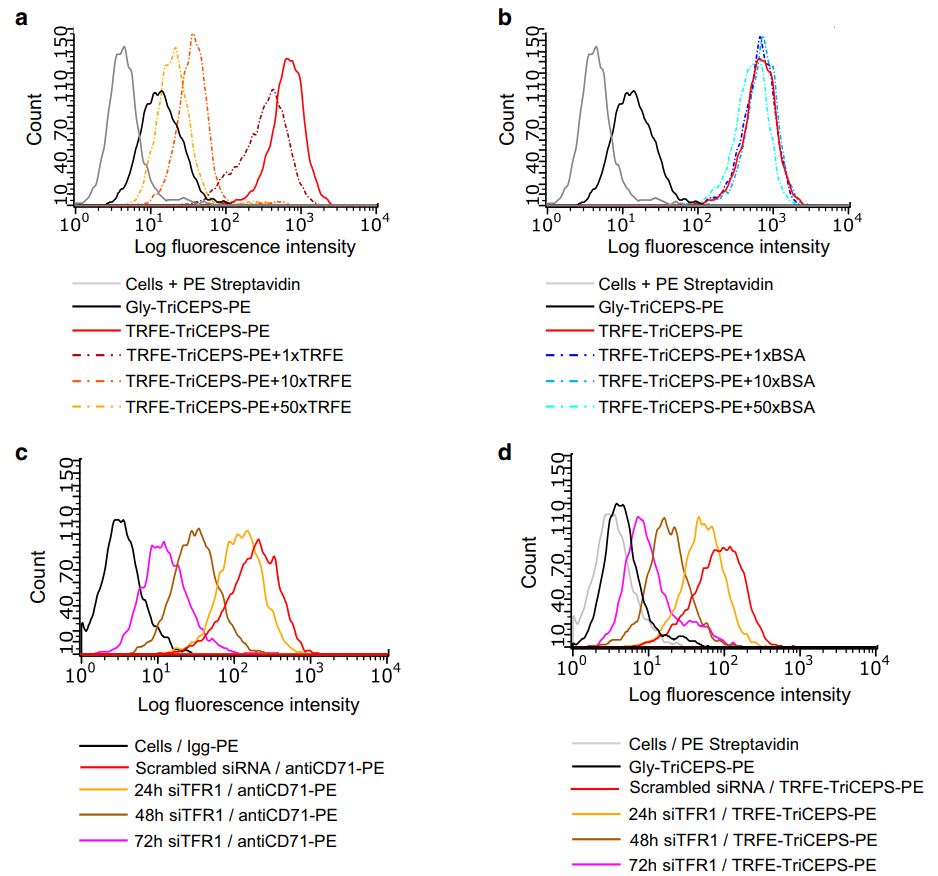
Lopez-Garcia LA, Demiray L, Ruch-Marder S, Hopp AK, Hottiger MO, Helbling PM, Pavlou MP
Validation of extracellular ligand-receptor interactions by Flow-TriCEPS
BMC Res Notes. 2018 Dec 5;11(1):863. doi: 10.1186/s13104-018-3974-5
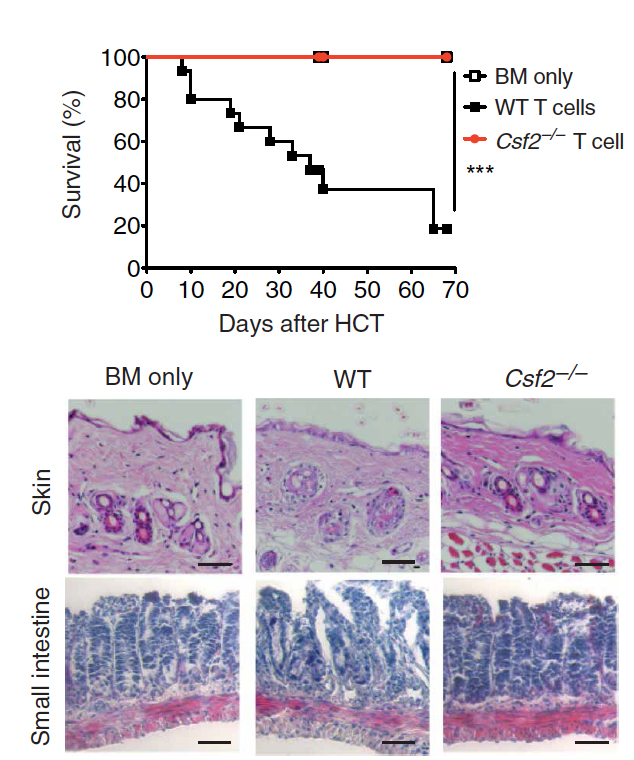
Tugues S, Amorim A, Spath S, Martin-Blondel G, Schreiner B, De Feo D, Lutz M, Guscetti F, Apostolova P, Haftmann C, Hasselblatt P, Núñez NG, Hottiger MO, van den Broek M, Manz MG, Zeiser R, Becher B
Graft-versus-host disease, but not graft-versus-leukemia immunity, is mediated by GM-CSF-licensed myeloid cells
Sci Transl Med. 2018 Nov 28;10(469). pii: eaat8410. doi: 10.1126/scitranslmed.aat8410
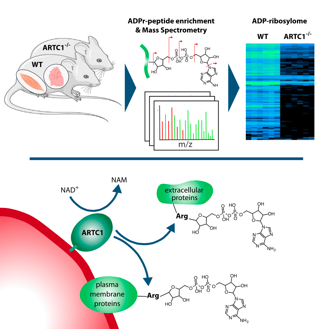
Leutert M, Menzel S, Braren R, Rissiek B, Hopp A, Nowak K, Bisceglie L, Gehrig P, Li H, Zolkiewska A, Koch-Nolte F, Hottiger MO
Proteomic Characterization of the Heart and Skeletal Muscle Reveals Widespread Arginine ADP-Ribosylation by the ARTC1 Ectoenzyme
Cell Rep. 2018 Aug 14;24(7):1916-1929.e5. doi: 10.1016/j.celrep.2018.07.048
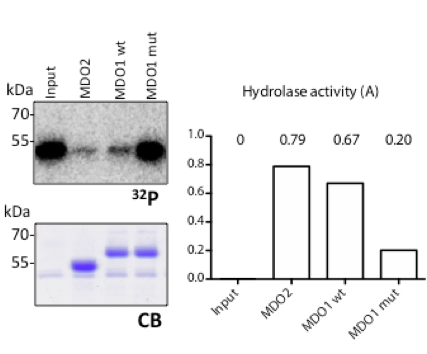
Abplanalp J, Hopp AK, Hottiger MO
Mono-ADP-Ribosylhydrolase Assays
Methods Mol Biol. 2018;1813:205-213. doi: 10.1007/978-1-4939-8588-3_13
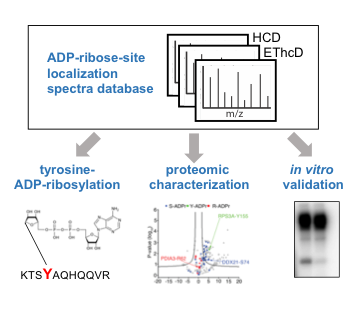
Leslie Pedrioli DM, Leutert M, Bilan V, Nowak K, Gunasekera K, Ferrari E, Imhof R, Malmström L, Hottiger MO
Comprehensive ADP-ribosylome analysis identifies tyrosine as an ADP-ribose acceptor site
EMBO Rep. 2018 Jun 28. pii: e45310
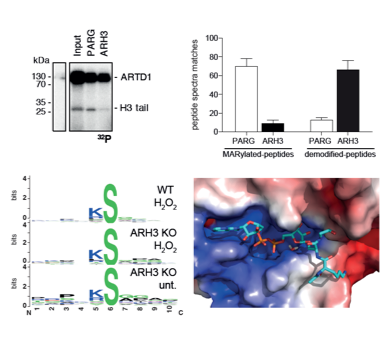
Abplanalp J, Leutert M, Frugier E, Nowak K, Feurer R, Kato J, Kistemaker HVA, Filippov DV, Moss J, Caflisch A, Hottiger MO
Proteomic analyses identify ARH3 as a serine mono-ADP-ribosylhydrolase
Nat Commun. 2017 Dec 12;8(1):2055
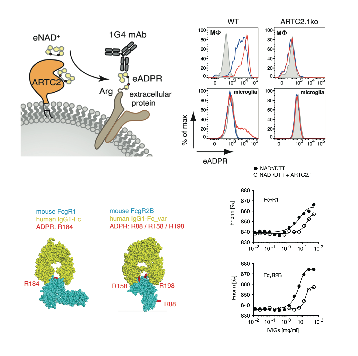
Rissiek B, Menzel S, Leutert M, Cordes M, Behr S, Jank L, Ludewig P, Gelderblom M, Rissiek A, Adriouch S, Haag F, Hottiger MO, Koch-Nolte F, Magnus T
Ecto-ADP-ribosyltransferase ARTC2.1 functionally modulates FcγR1 and FcγR2B on murine microglia
Sci Rep. 2017 Nov 28;7(1):16477
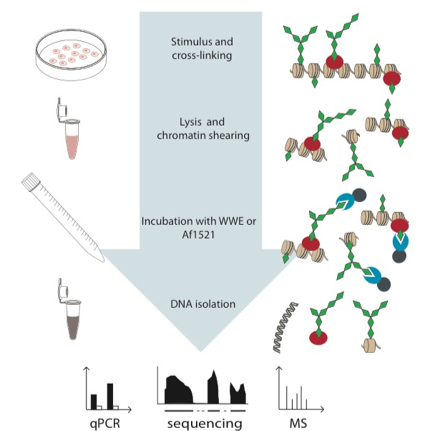
Bisceglie L, Bartolomei G, Hottiger MO
ADP-ribose-specific chromatin-affinity purification for investigating genome-wide or locus-specific chromatin ADP-ribosylation
Nat Protoc. 2017 Sep;12(9):1951-1961
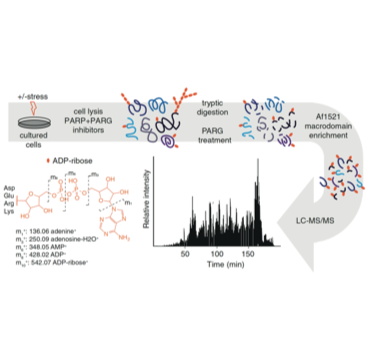
Larsen SC, Leutert M, Bilan V, Martello R, Jungmichel S, Young C, Hottiger MO, Nielsen ML
Proteome-Wide Identification of In Vivo ADP-Ribose Acceptor Sites by Liquid Chromatography-Tandem Mass Spectrometry
Methods Mol Biol. 2017;1608:149-162
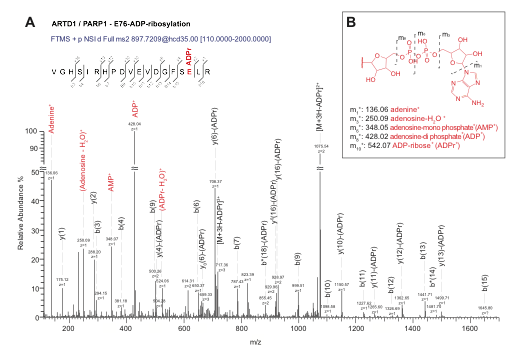
Leutert M, Bilan V, Gehrig P, Hottiger MO
Identification of ADP-Ribose Acceptor Sites on In Vitro Modified Proteins by Liquid Chromatography-Tandem Mass Spectrometry
Methods Mol Biol. 2017;1608:137-148
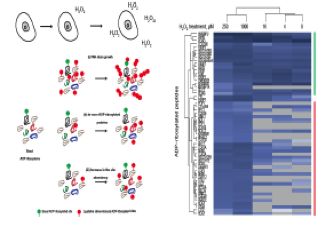
Bilan V, Selevsek N, Kistemaker HA, Abplanalp J, Feurer R, Filippov DV & Hottiger MO
New quantitative mass spectrometry approaches reveal different ADP-ribosylation phases dependent on the levels of oxidative stress
Mol. Cell. Proteomics 16(5):949 (2017)
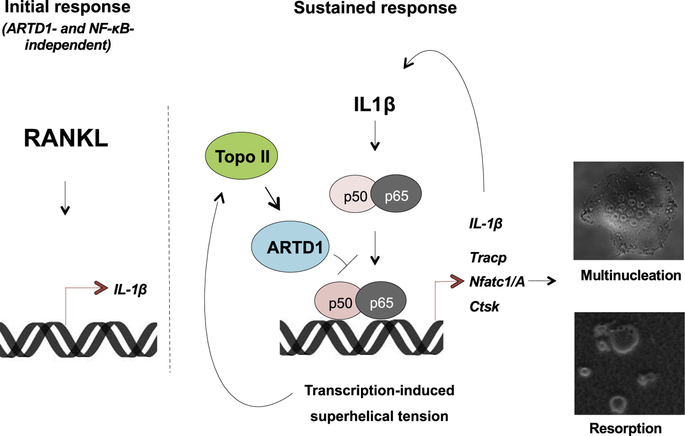
Robaszkiewicz A, Qu C, Wisnik E, Ploszaj T, Mirsaidi A, Kunze FA, Richards PJ, Cinelli P, Mbalaviele G, Hottiger MO
ARTD1 regulates osteoclastogenesis and bone homeostasis by dampening NF-κB-dependent transcription of IL-1β
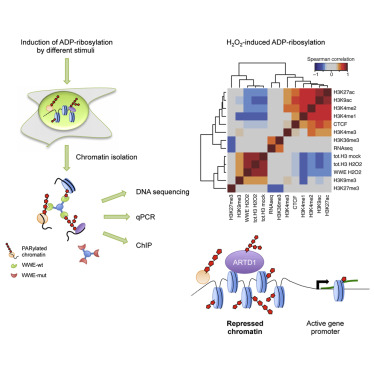
Bartolomei G, Leutert M, Manzo M, Baubec T, Hottiger MO
Analysis of Chromatin ADP-Ribosylation at the Genome-wide Level and at Specific Loci by ADPr-ChAP
Mol Cell. 2016 Feb 4;61(3):474-85
Highlighted in ADPr-ChAP: Mapping ADP-Ribosylation onto the Genome. Mol Cell. 2016;61(3):327-8
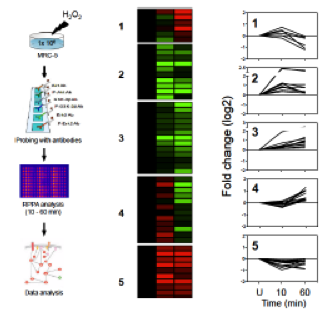
Andersson A, Bluwstein A, Kumar N, Teloni F, Traenkle J, Baudis M, Altmeyer M & Hottiger MO
PKCalpha and HMGB1 antagonistically control hydrogen peroxide-induced poly-ADP-ribose formation
Nucleic Acids Res. 44(16):7630 (2016)
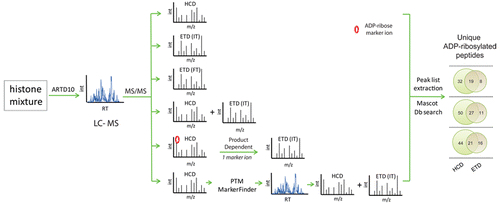
Rosenthal F, Nanni P, Barkow-Oesterreicher S, Hottiger MO
Optimization of LTQ-Orbitrap Mass Spectrometer Parameters for the Identification of ADP-Ribosylation Sites
J Proteome Res. 2015 Sep 4;14(9):4072-9
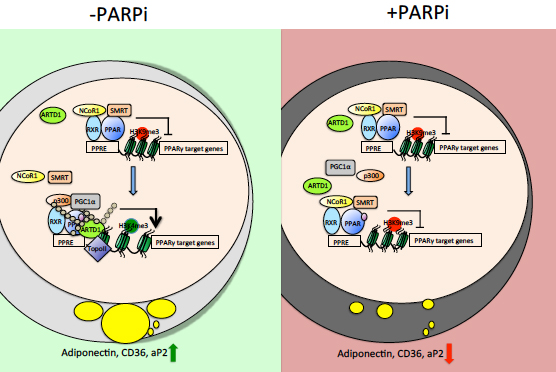
Lehmann M, Pirinen E, Mirsaidi A, Kunze FA, Richards PJ, Auwerx J, Hottiger MO
ARTD1-induced poly-ADP-ribose formation enhances PPARγ ligand binding and co-factor exchange
Nucleic Acids Res. 2015 Jan;43(1):129-42
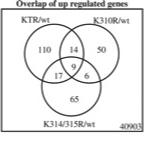
Weber FA, Bartolomei G, Hottiger MO, Cinelli P
Artd1/Parp1 regulates reprogramming by transcriptional regulation of Fgf4 via Sox2 ADP-ribosylation
Stem Cells. 2013 Nov;31(11):2364-73
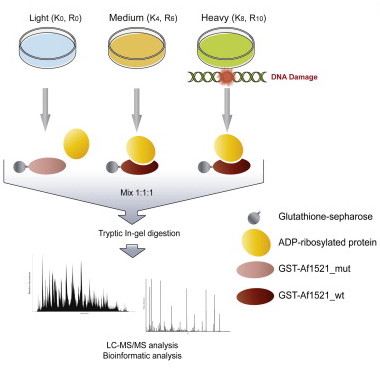
Jungmichel S, Rosenthal F, Altmeyer M, Lukas J, Hottiger MO, Nielsen ML
Proteome-wide identification of poly(ADP-Ribosyl)ation targets in different genotoxic stress responses
Mol Cell. 2013 Oct 24;52(2):272-85
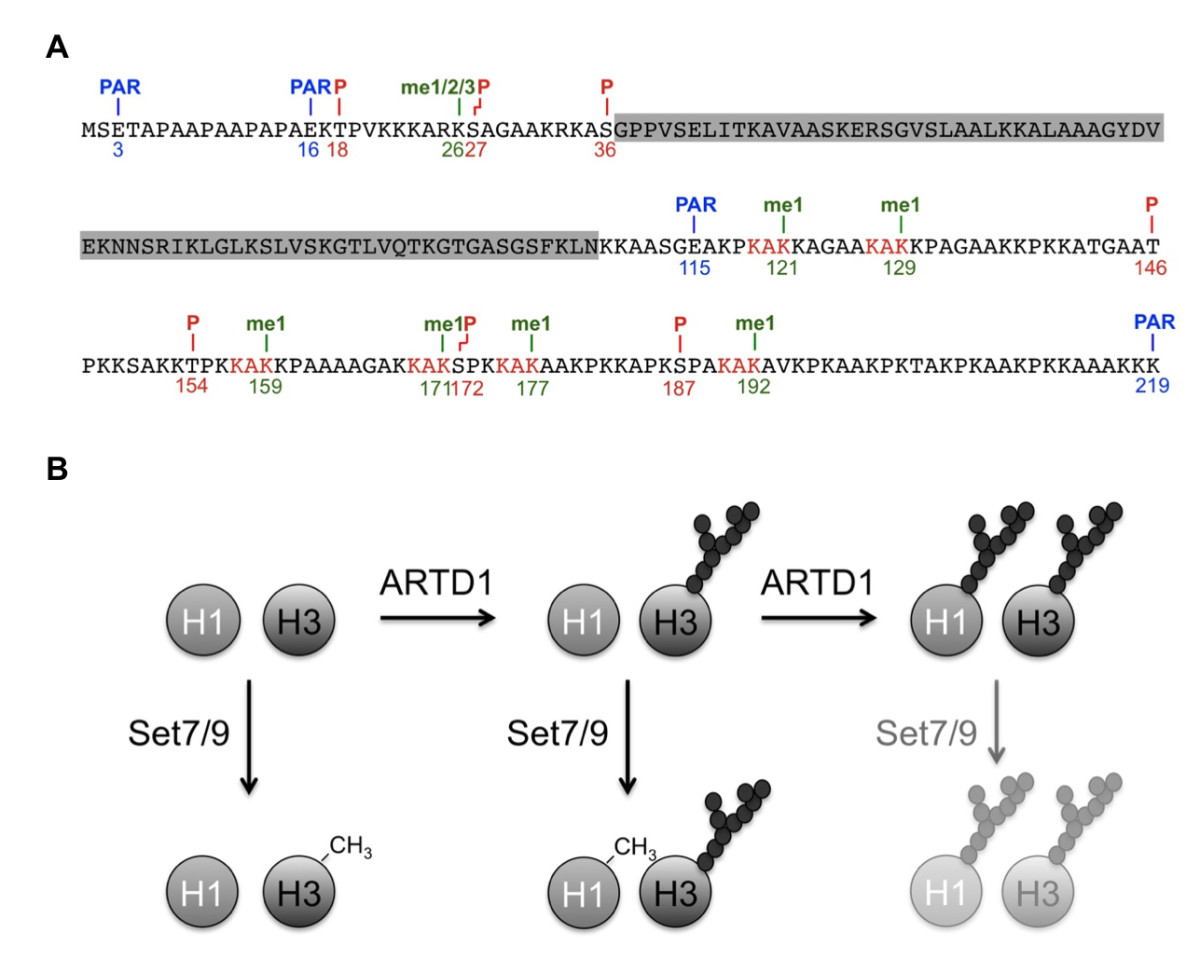
Kassner I, Barandun M, Fey M, Rosenthal F, Hottiger MO
Crosstalk between SET7/9-dependent methylation and ARTD1-mediated ADP-ribosylation of histone H1.4
Epigenetics Chromatin. 2013 Jan 5;6(1):1
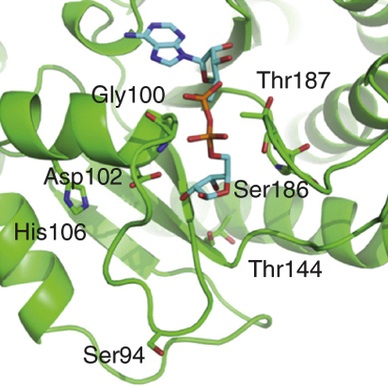
Rosenthal F, Feijs KL, Frugier E, Bonalli M, Forst AH, Imhof R, Winkler HC, Fischer D, Caflisch A, Hassa PO, Lüscher B, Hottiger MO
Macrodomain-containing proteins are novel mono-ADP-ribosylhydrolases
Nat Struct Mol Biol 2013;20(4):502-7

Erener S, Pétrilli V, Kassner I, Minotti R, Castillo R, Santoro R, Hassa PO, Tschopp J, Hottiger M
Inflammasome-Activated Caspase 7 Cleaves PARP1 to Enhance the Expression of a Subset of NF-κB Target Genes
Mol Cell 2012 Apr 27;46(2):200-11
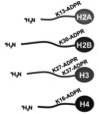
Messner S, Altmeyer M, Zhao H, Pozivil A, Roschitzki B, Gehrig P, Rutishauser D, Huang D, Caflisch A, Hottiger MO
PARP1 ADP-ribosylates lysine residues of the core histone tails
Nucleic Acids Res 2010;38(19):6350-6362