Publications - Prof. Dr. Matthias Altmeyer
Full list of publications
Featured publications
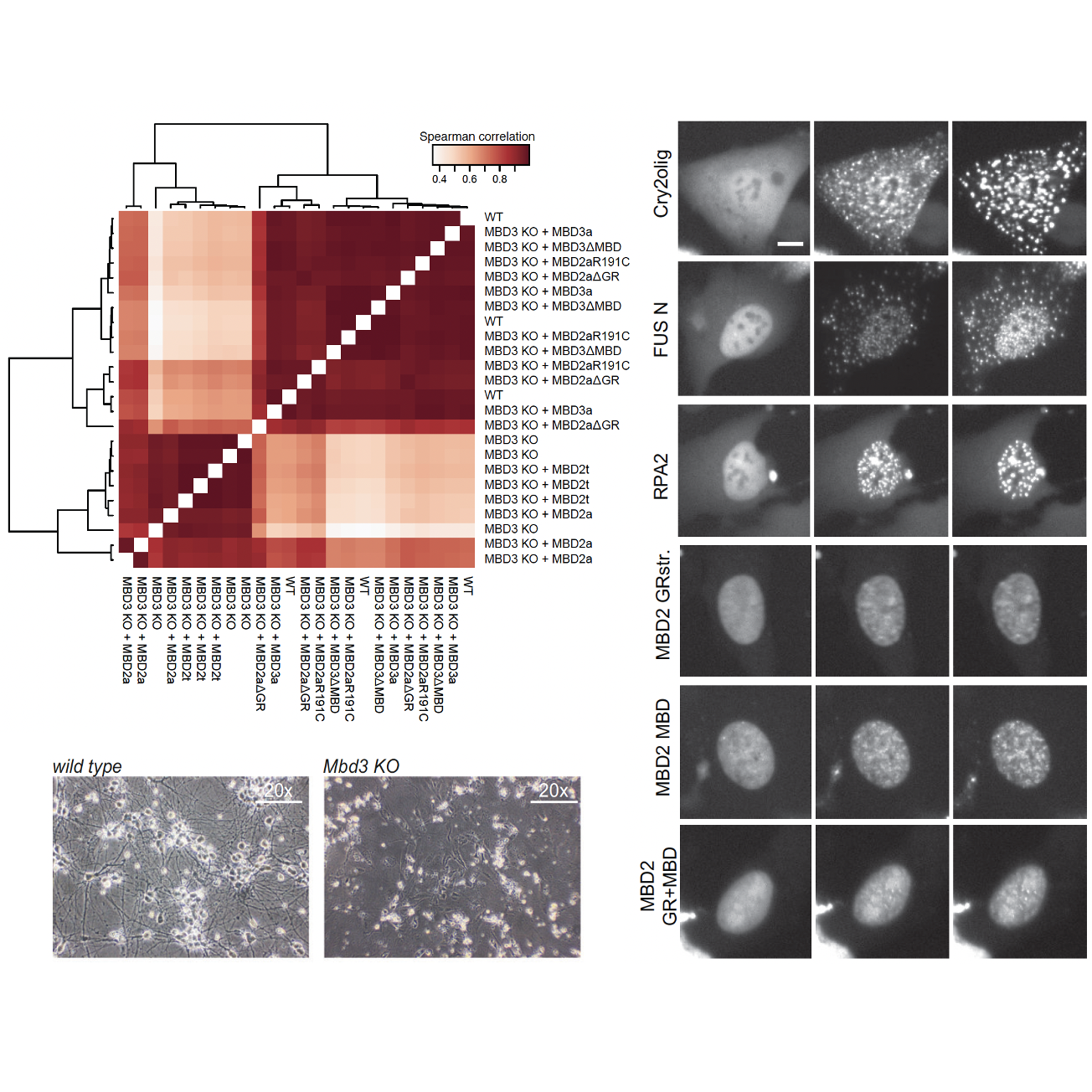
Nina Schmolka, Ino D Karemaker, Richard Cardoso da Silva, Davide C Recchia, Vincent Spegg, Jahnavi Bhaskaran, Michael Teske, Nathalie P de Wagenaar, Matthias Altmeyer, Tuncay Baubec
Nat Commun. 2023 Jun 29;14(1):3848.
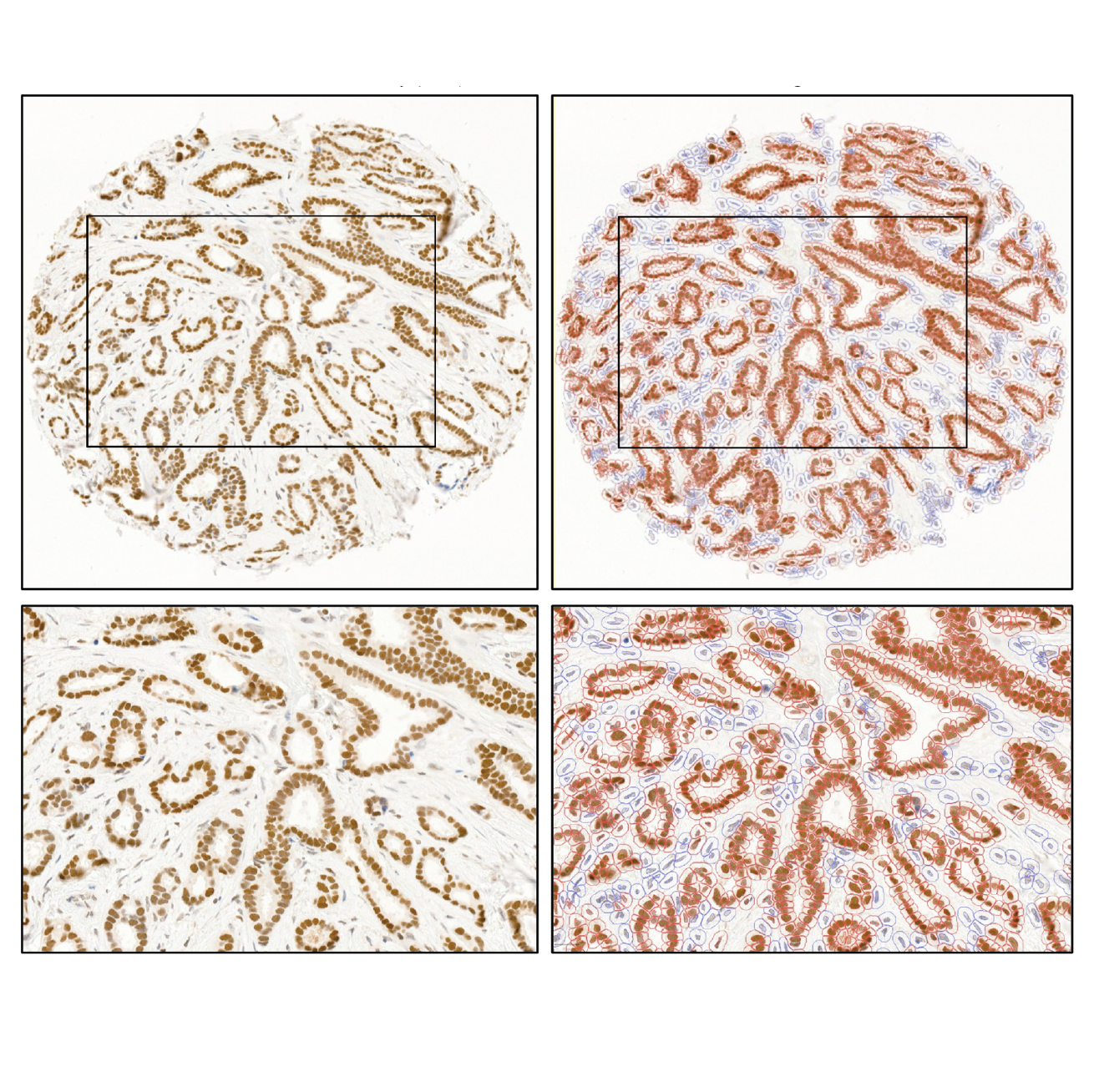
Aswini Krishnan, Vincent Spegg, Susanne Dettwiler, Peter Schraml, Holger Moch, Konstantin Dedes, Zsuzsanna Varga, Matthias Altmeyer
Mod Pathol. 2023 Mar 27;36(7):100167.

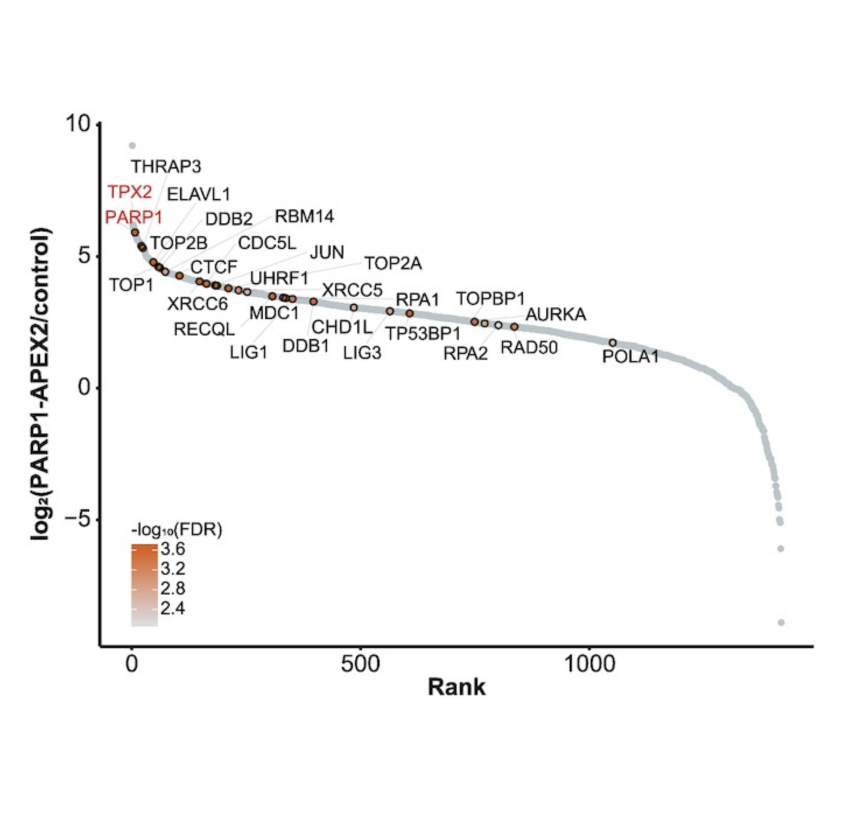
Vincent Spegg, Andreas Panagopoulos, Merula Stout, Aswini Krishnan, Giordano Reginato, Ralph Imhof, Bernd Roschitzki, Petr Cejka, Matthias Altmeyer
Nat Struct Mol Biol. 2023 Apr;30(4):451-462.
Thorsten Mosler, H Irem Baymaz, Justus F Gräf, Ivan Mikicic, Georges Blattner, Edward Bartlett, Matthias Ostermaier, Rossana Piccinno, Jiwen Yang, Andrea Voigt, Marco Gatti, Stefania Pellegrino, Matthias Altmeyer, Katja Luck, Ivan Ahel, Vassilis Roukos, Petra Beli
Nucleic Acids Res. 2022 Nov 11;50(20):11600-11618.
Singh JK, Noordermeer SM, Jimenez-Sainz J, Maranon DG, Altmeyer M
Front Genet. 2022 Aug 12;13:993889.
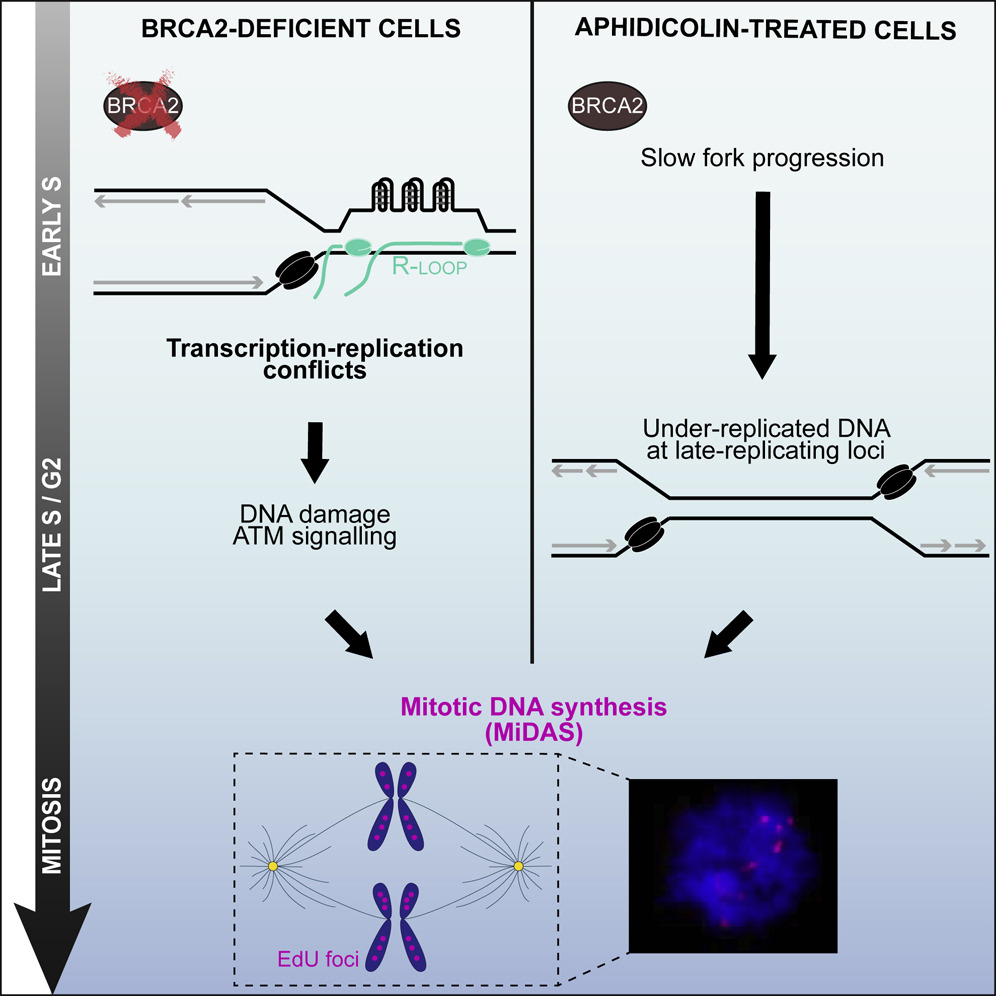
Groelly FJ, Dagg RA, Petropoulos M, Rossetti GG, Prasad B, Panagopoulos A, Paulsen T, Karamichali A, Jones SE, Ochs F, Dionellis VS, Puig Lombardi E, Miossec MJ, Lockstone H, Legub e G, Blackford AN, Altmeyer M, Halazonetis TD, Tarsounas M
Mitotic DNA synthesis is caused by transcription-replication conflicts in BRCA2-deficient cells
Mol Cell. 2022 Aug 17:S1097-2765(22)00707-9.

Bolck HA, Przetocka S, Meier R, von Aesch C, Zurfluh C, Hänggi K, Spegg V, Altmeyer M, Stebler M, Nørrelykke SF, Horvath P, Sartori AA, Porro A
RNAi Screening Uncovers a Synthetic Sick Interaction between CtIP and the BARD1 Tumor Suppressor
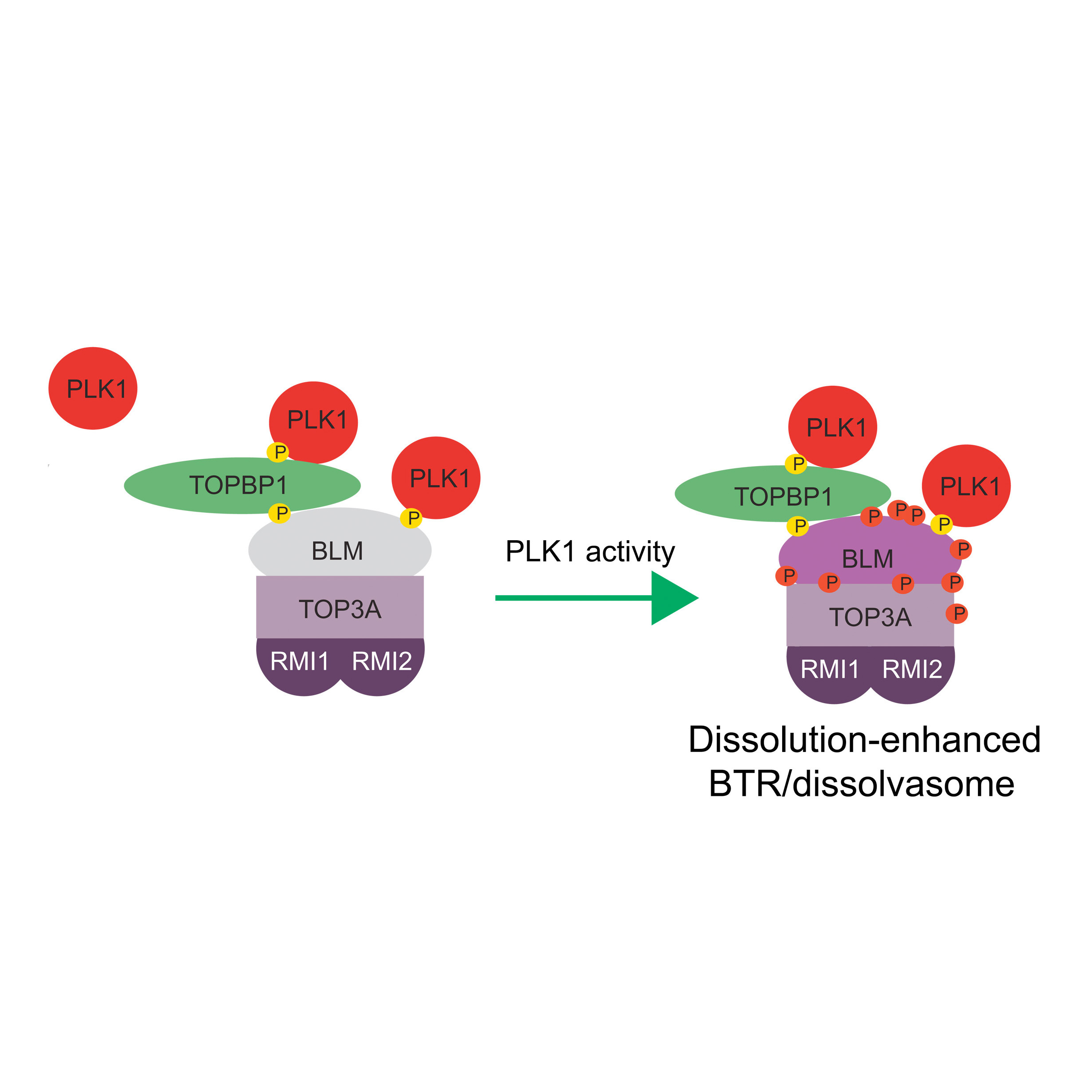
Balbo Pogliano C, Ceppi I, Giovannini S, Petroulaki V, Palmer N, Uliana F, Gatti M, Kasaciunaite K, Freire R, Seidel R, Altmeyer M, Cejka P, Matos J
The CDK1-TOPBP1-PLK1 axis regulates the Bloom's syndrome helicase BLM to suppress crossover recombination in somatic cells
Sci Adv. 2022 Feb 4;8(5):eabk0221.
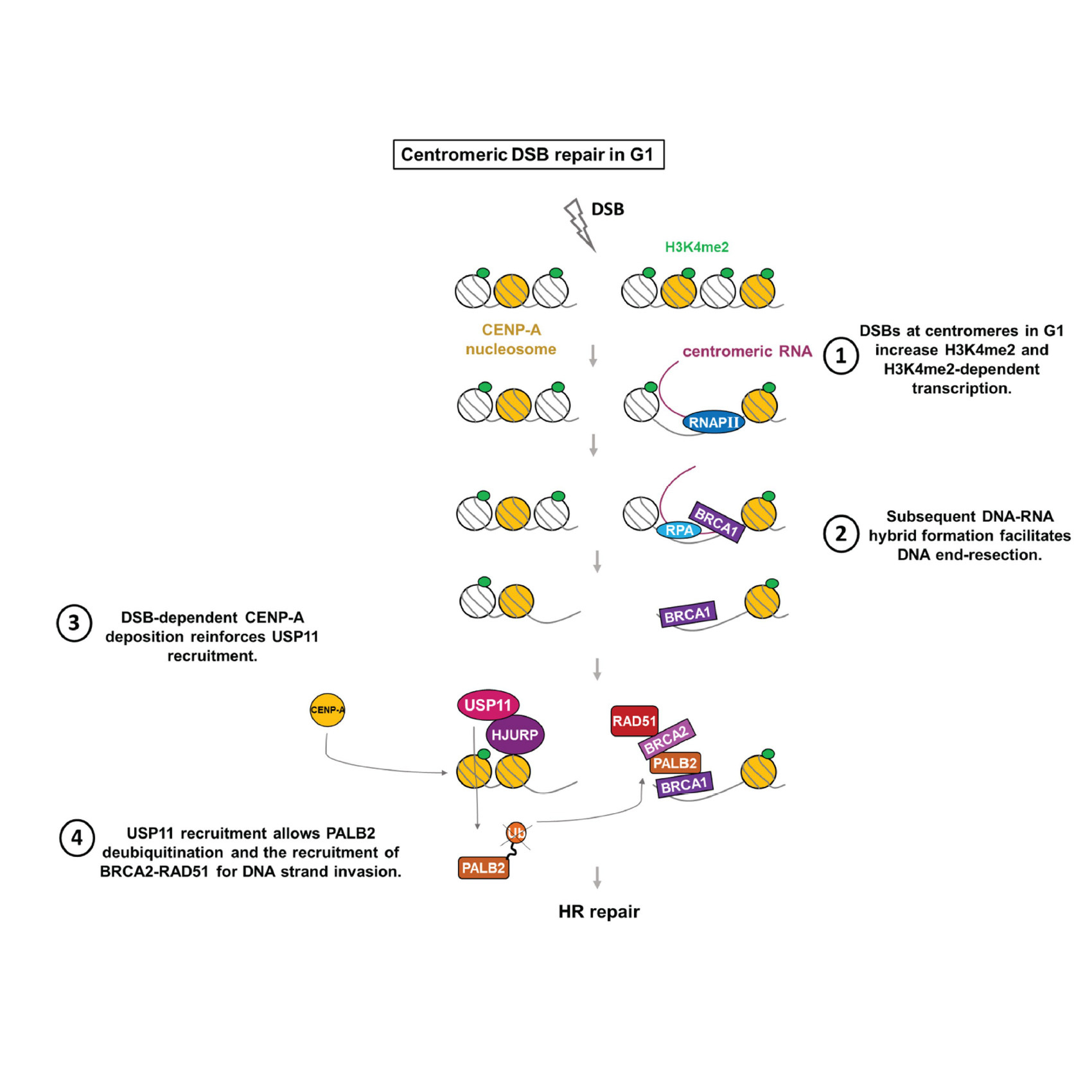
Yilmaz D, Furst A, Meaburn K, Lezaja A, Wen Y, Altmeyer M, Reina-San-Martin B, Soutoglou E
Activation of homologous recombination in G1 preserves centromeric integrity
Nature. 2021 Dec;600(7890):748-753.
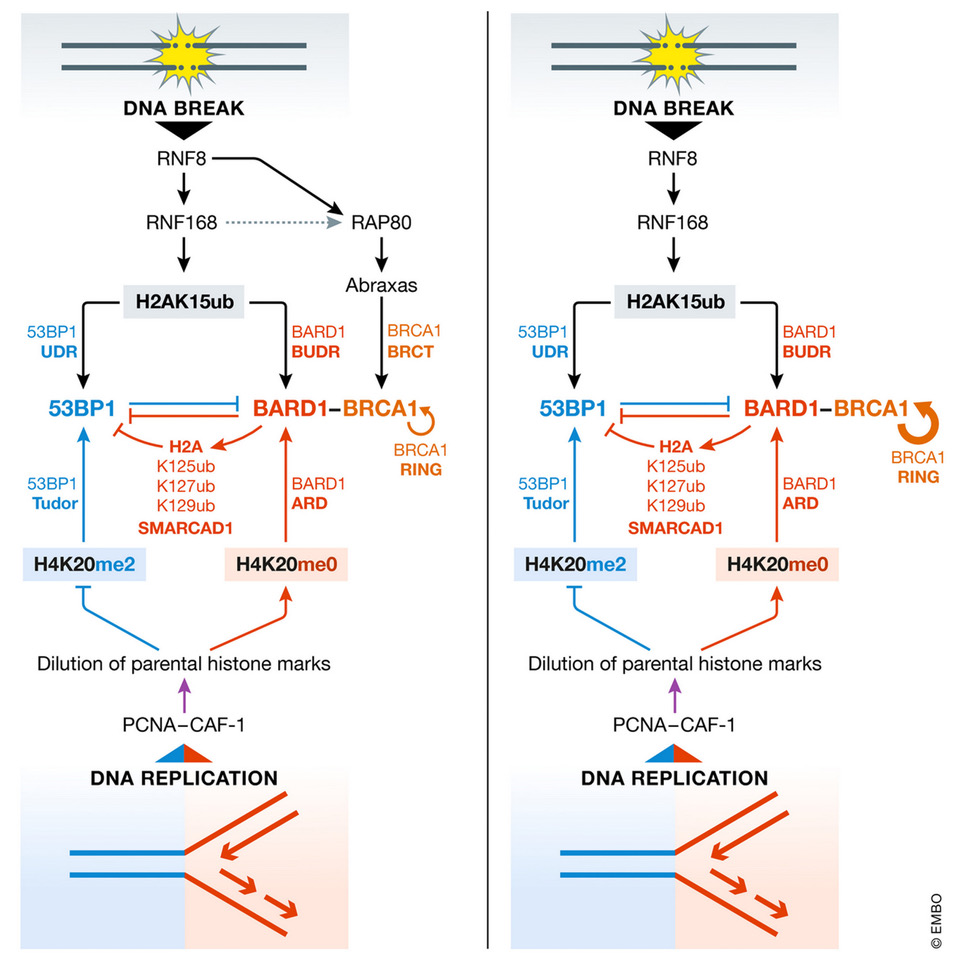
Andreas Panagopoulos, Matthias Altmeyer
When the RAP (80) fades out, you can hear BRCA1 RING

Vincent Spegg, Matthias Altmeyer
Biomolecular condensates at sites of DNA damage: More than just a phase
DNA Repair (Amst). 2021 Oct;106:103179.
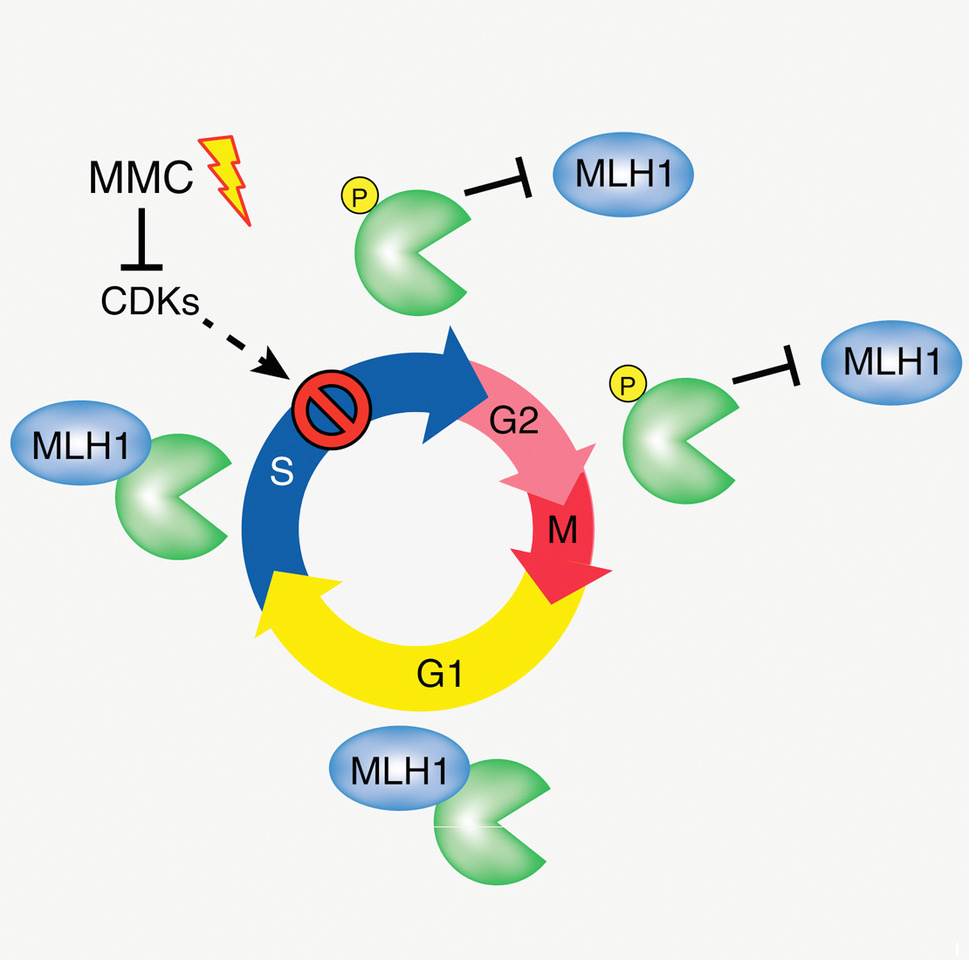
Porro A, Mohiuddin M, Zurfluh C, Spegg V, Dai J, Iehl F, Ropars V, Collotta G, Fishwick KM, Mozaffari NL, Guérois R, Jiricny J, Altmeyer M, Charbonnier JB, Pearson CE, Sartori AA
FAN1-MLH1 interaction affects repair of DNA interstrand cross-links and slipped-CAG/CTG repeats
Sci Adv. 2021 Jul 30;7(31):eabf7906.

Lüscher B, Ahel I, Altmeyer M et al.
ADP-ribosyltransferases, an update on function and nomenclature
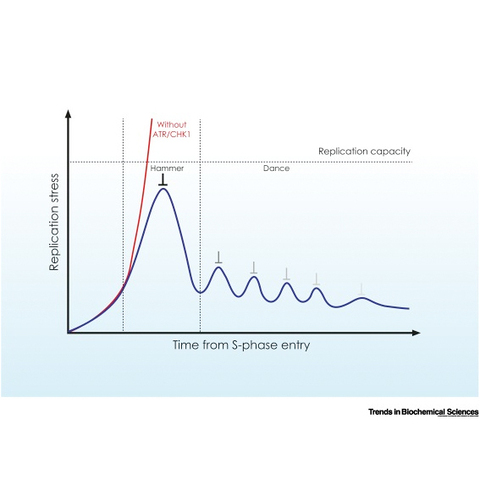
Panagopoulos A, Altmeyer M
The Hammer and the Dance of Cell Cycle Control
Trends Biochem Sci. 2021 Apr;46(4):301-314.
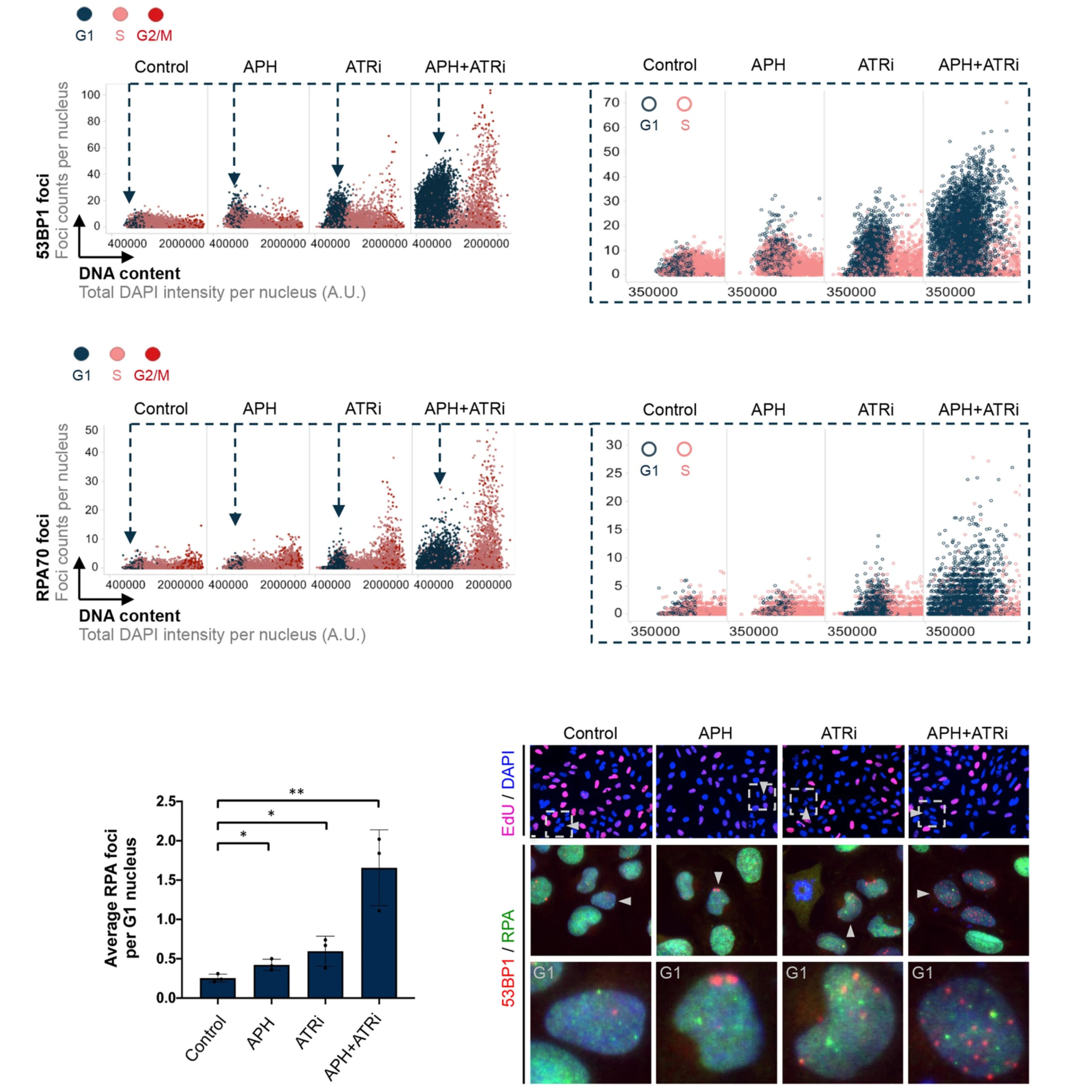
Lezaja A, Panagopoulos A, Wen Y, Carvalho E, Imhof R, Altmeyer M
RPA shields inherited DNA lesions for post-mitotic DNA synthesis
Nat Commun. 2021 Jun 22;12(1):3827
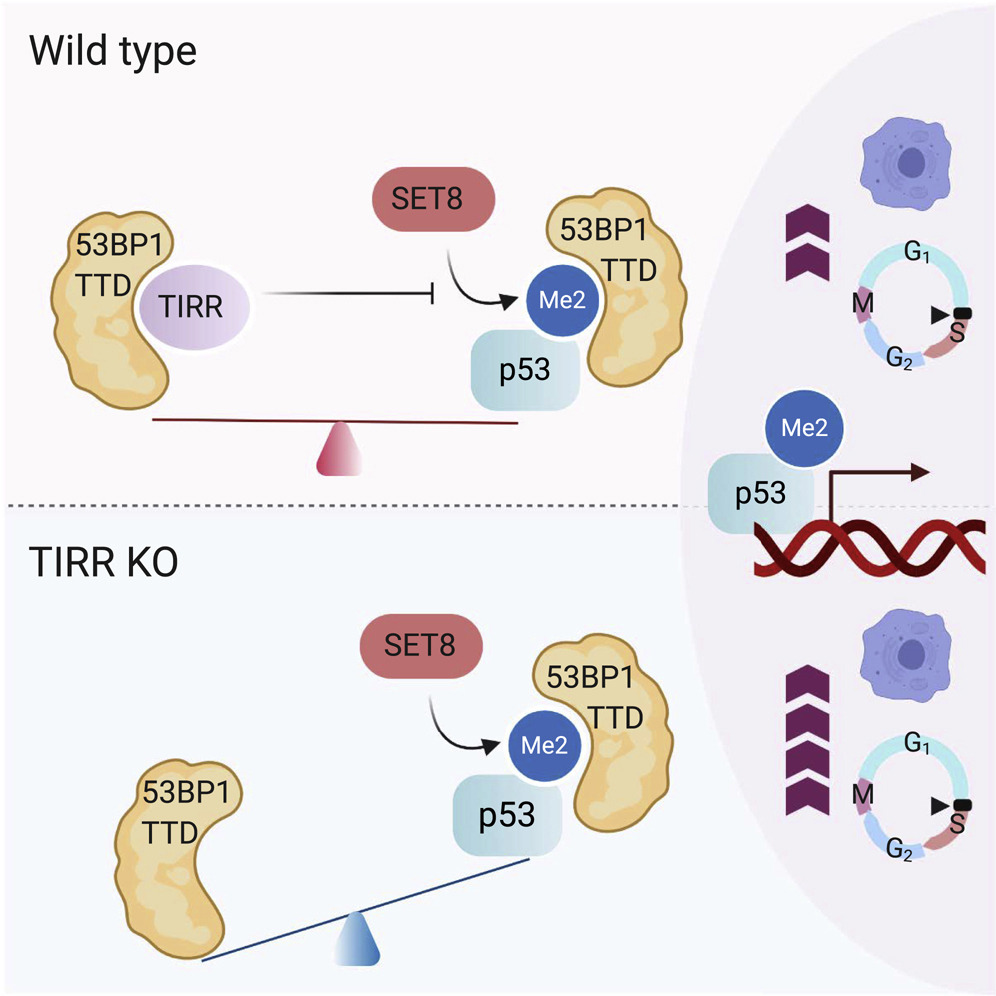
Parnandi N, Rendo V, Cui G, Botuyan MV, Remisova M, Nguyen H, Drané P, Beroukhim R, Altmeyer M, Mer G, Chowdhury D
TIRR inhibits the 53BP1-p53 complex to alter cell-fate programs
Mol Cell. 2021 Jun 17;81(12):2583-2595.e6.
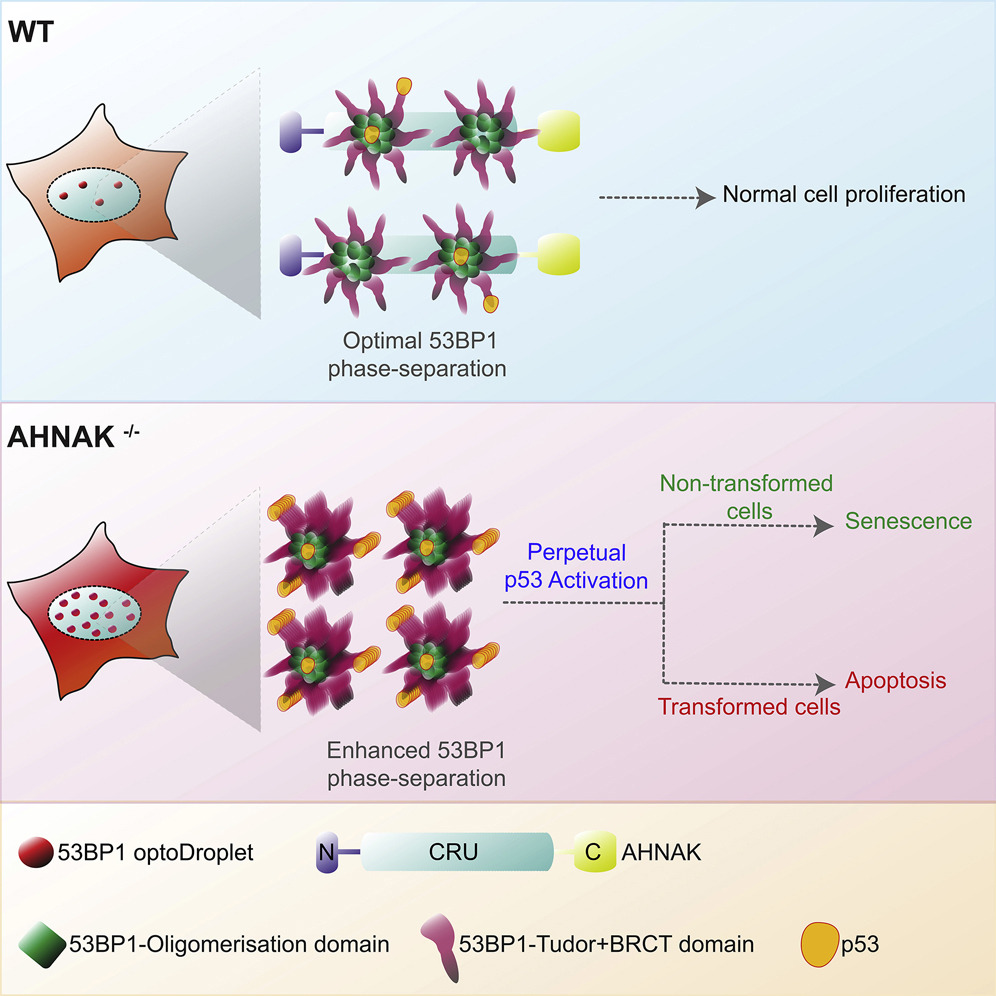
Ghodke I, Remisova M, Furst A, Kilic S, Reina-San-Martin B, Poetsch AR, Altmeyer M, Soutoglou E
AHNAK controls 53BP1-mediated p53 response by restraining 53BP1 oligomerization and phase separation
Mol Cell. 2021 Jun 17;81(12):2596-2610.e7.
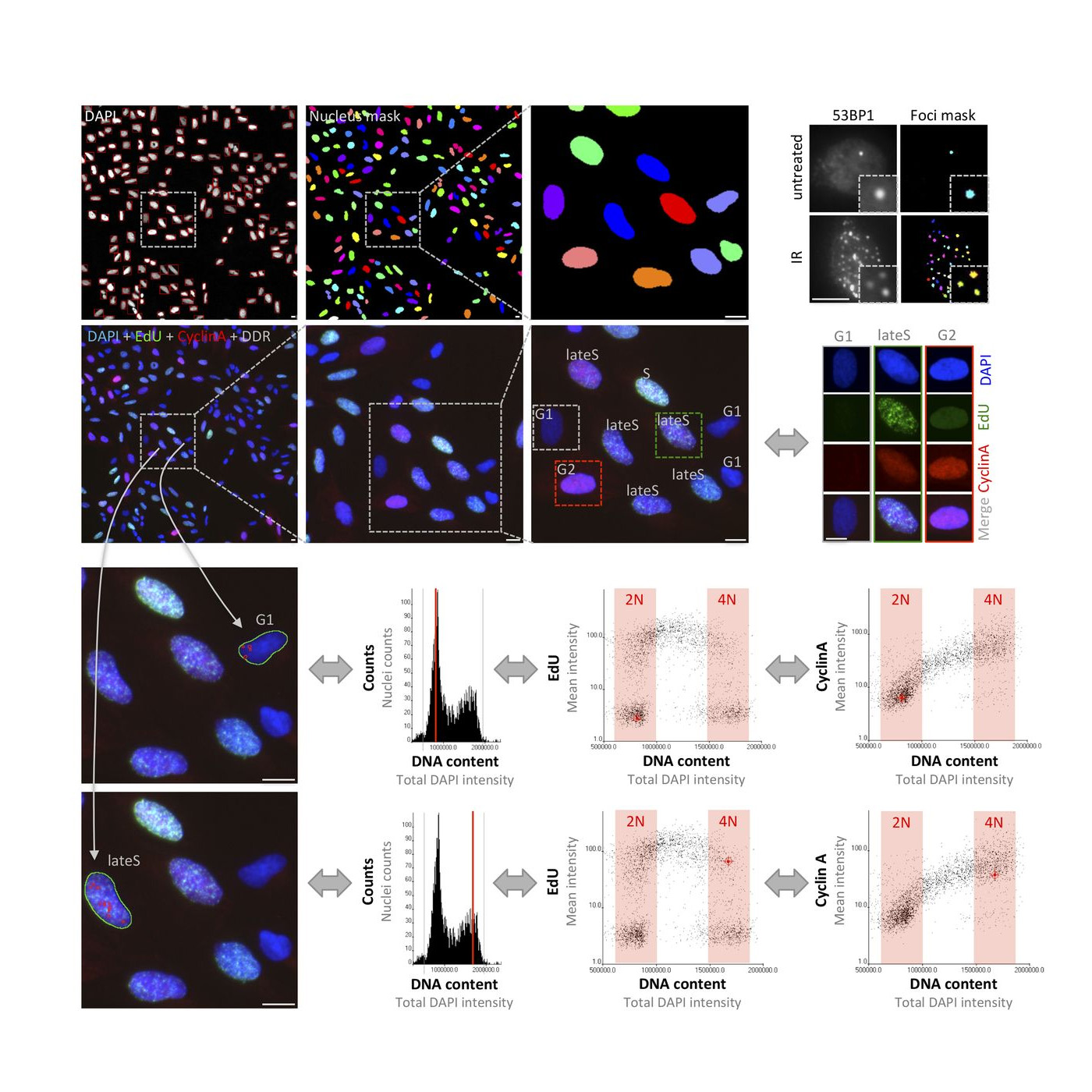
Michelena J, Pellegrino S, Spegg V, Altmeyer M
Replicated chromatin curtails 53BP1 recruitment in BRCA1-proficient and BRCA1-deficient cells
Life Sci Alliance. 2021 Apr 2;4(6):e202101023.
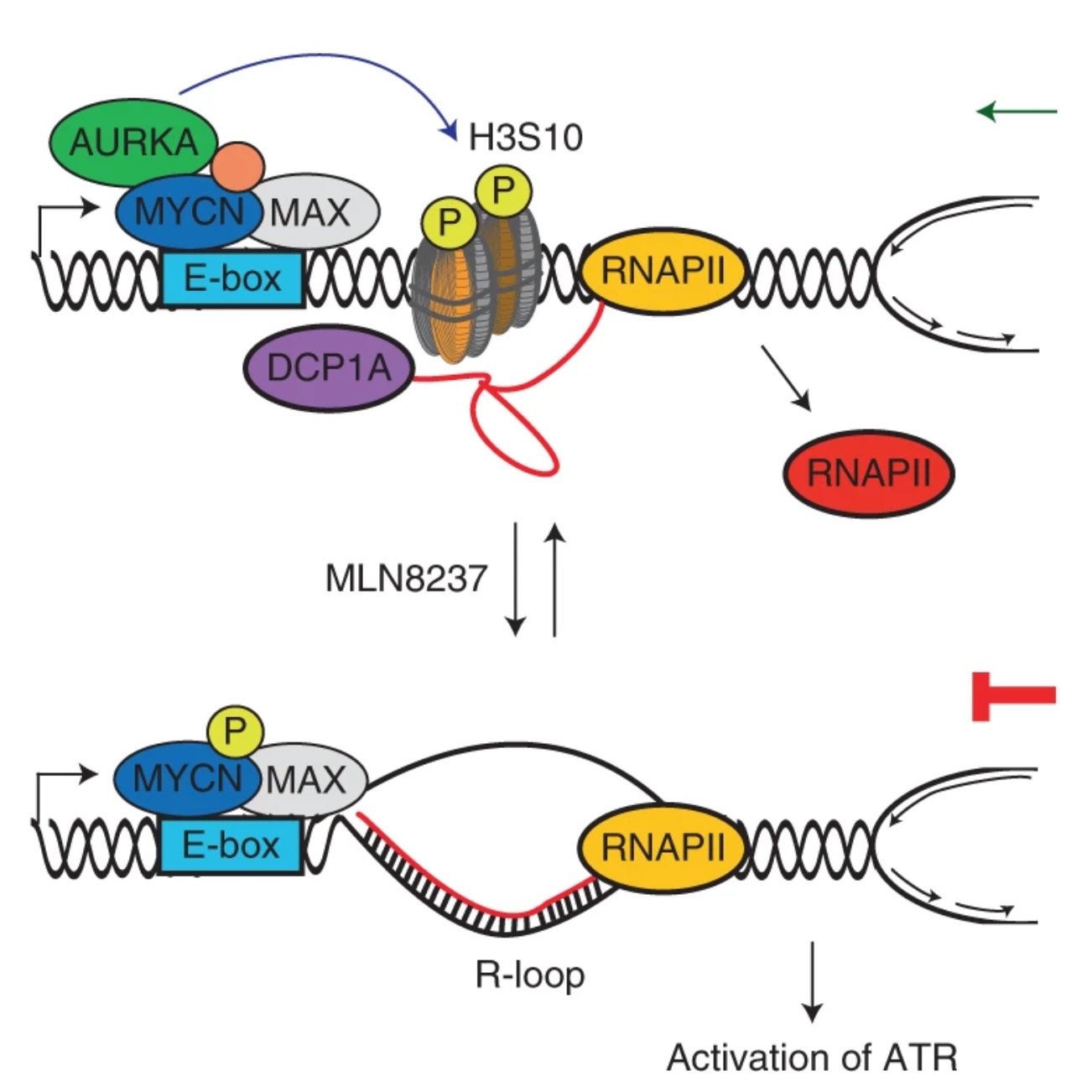
Roeschert I, Poon E, Henssen AG, Garcia HD, Gatti M, Giansanti C, Jamin Y, Ade CP, Gallant P, Schülein-Völk C, Beli P, Richards M, Rosenfeldt M, Altmeyer M, Anderson J, Eggert A, Dobbelstein M, Bayliss R, Chesler L, Büchel G, Eilers M
Combined inhibition of Aurora-A and ATR kinase results in regression of MYCN-amplified neuroblastoma
Nat Cancer. 2021 Mar;2(3):312-326.
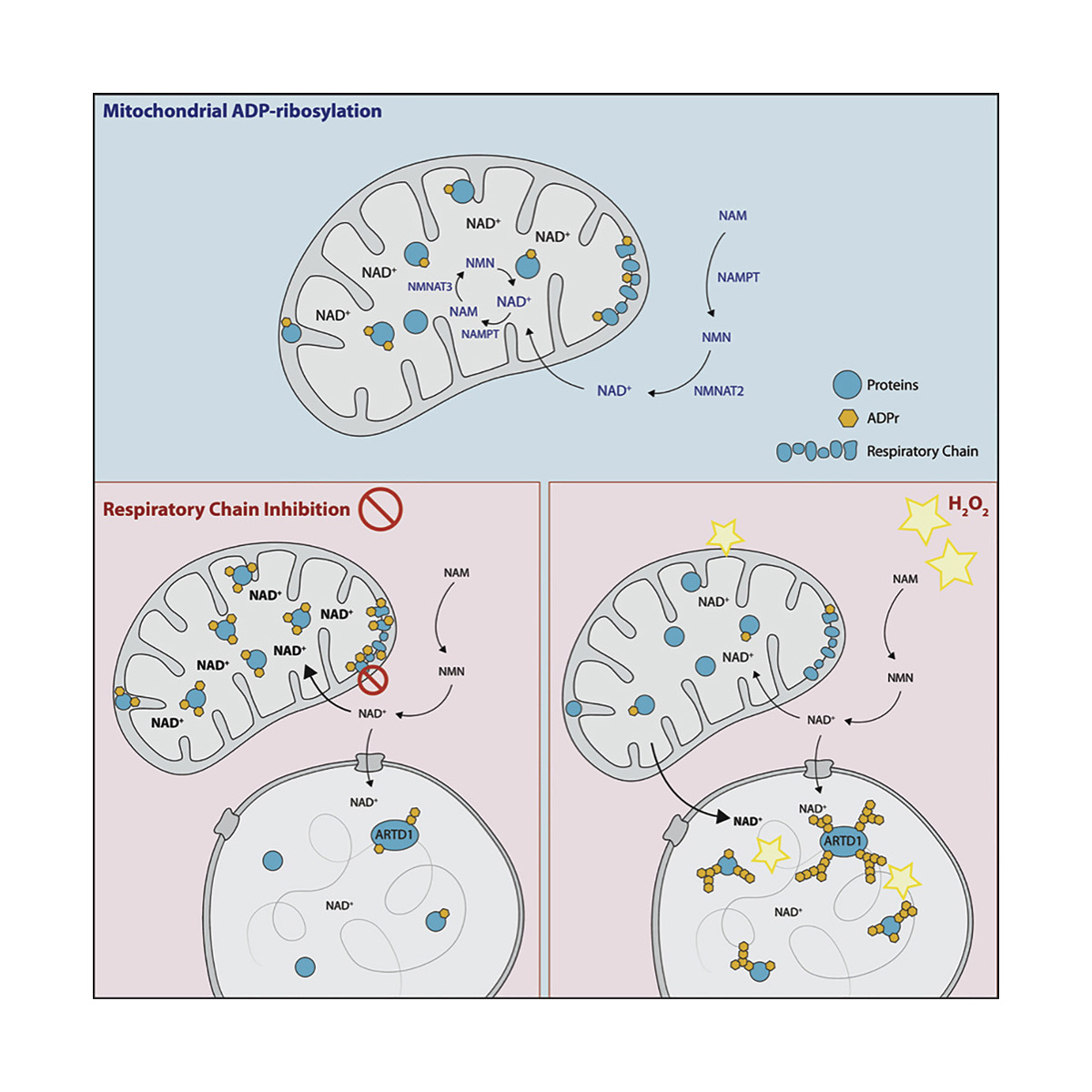
Hopp AK, Teloni F, Bisceglie L, Gondrand C, Raith F, Nowak K, Muskalla L, Howald A, Pedrioli PGA, Johnsson K, Altmeyer M, Pedrioli DML, Hottiger MO
Mitochondrial NAD+ Controls Nuclear ARTD1-Induced ADP-Ribosylation
Mol Cell. 2021 Jan 21;81(2):340-354.e5.
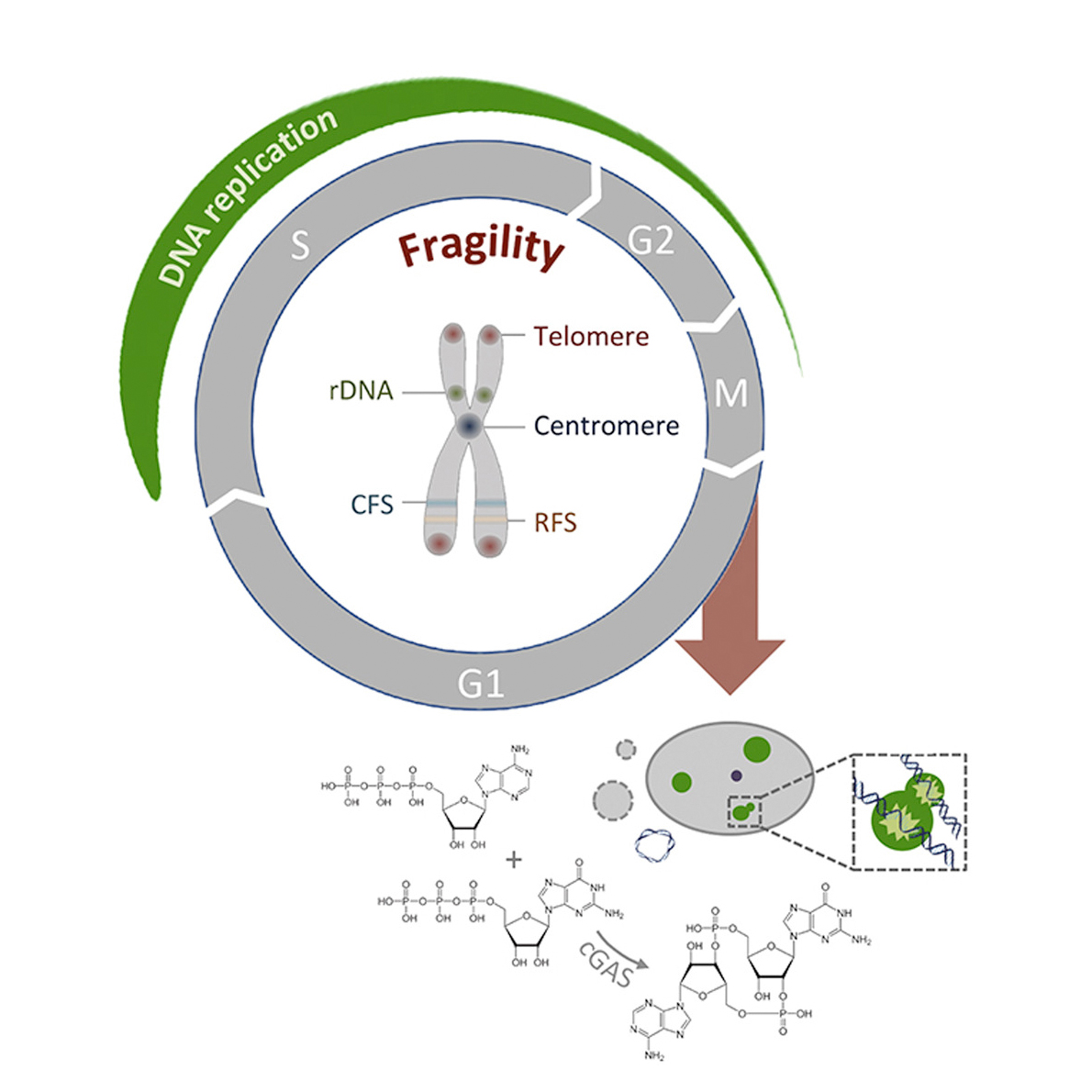
Lezaja A and Altmeyer M
Dealing with DNA lesions: When one cell cycle is not
enough
Curr Opin Cell Biol 2020 Dec 10;70:27-36
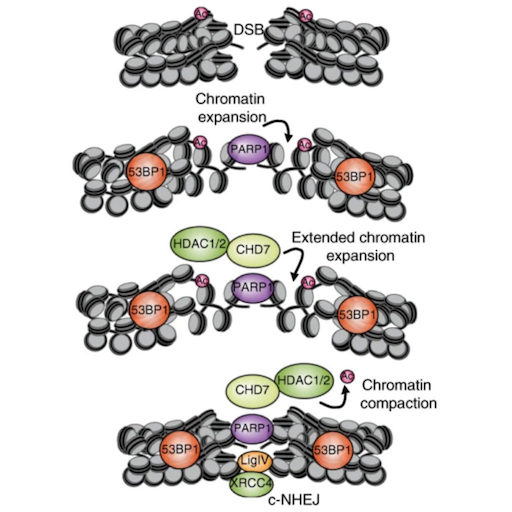
Rother M.B*, Pellegrino S*, Smith R, Gatti M, Meisenberg C, Wiegant W.W, Luijsterburg M.S, Imhof R, Downs J.A, Vertegaal A.C.O, Huet S, Altmeyer M & van Attikum H
* equal contribution
CHD7 and 53BP1 regulate distinct pathways for the re-ligation of DNA double-strand breaks
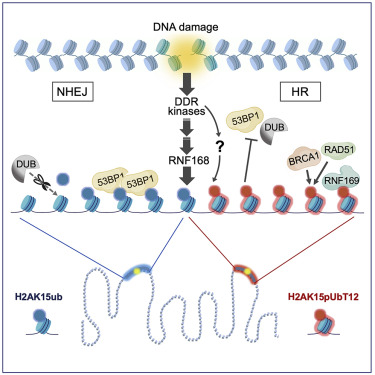
Walser F, Mulder M, Bragantini B, Burger S, Gubser T, Gatti M, Botuyan M.V, Villa A, Altmeyer M, Neri D, Ovaa H, Mer G and Penengo L
Ubiquitin Phosphorylation at Thr12 Modulates the DNA Damage Response
2020 Sep 29;S1097-2765(20)30647-X
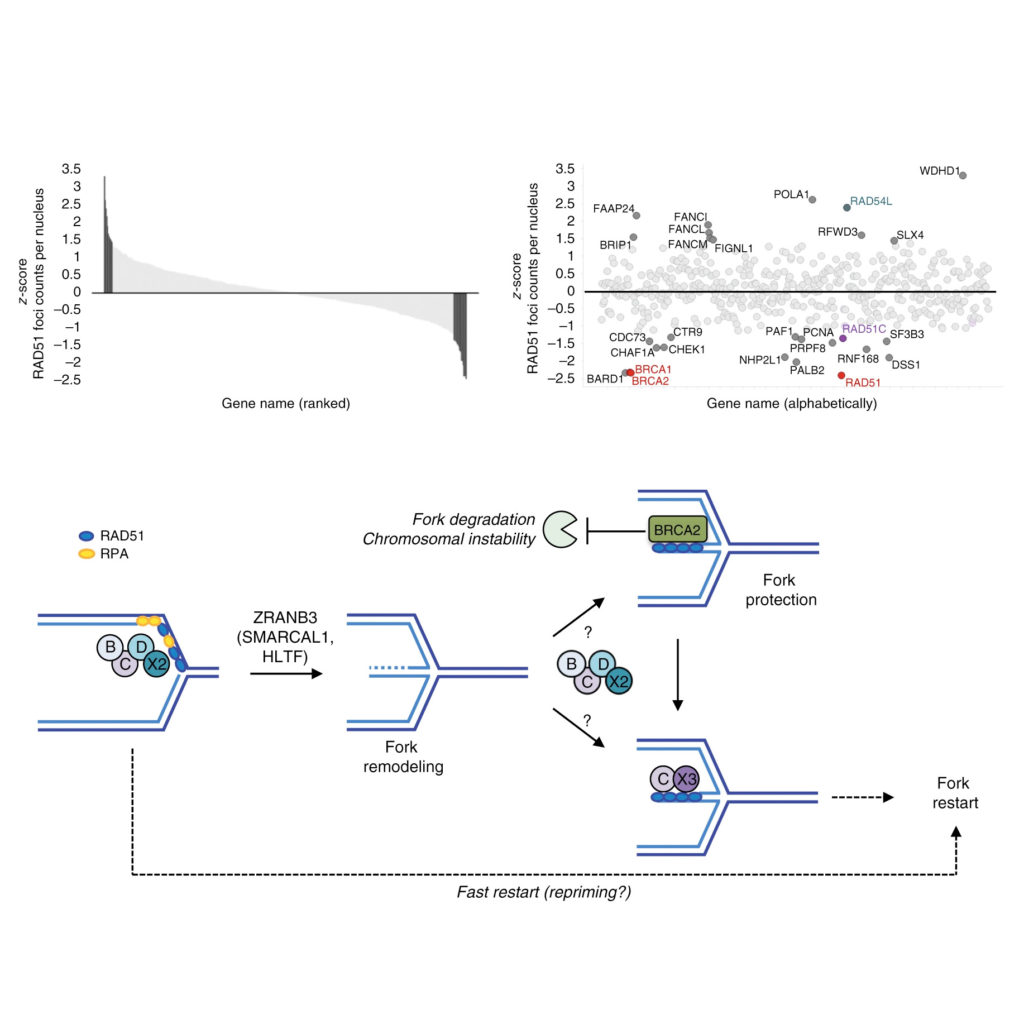
Berti M*, Teloni F*, Mijic S, Ursich S, Fuchs J, Palumbieri M.D, Krietsch J, Schmid J.A, Garcin E.B, Gon S, Modesti M, Altmeyer M" & Lopes M"
* equal contribution
Sequential role of RAD51 paralog complexes in replication fork remodeling and restart
Nature Communications 2020 Jul 15;11(1):3531.
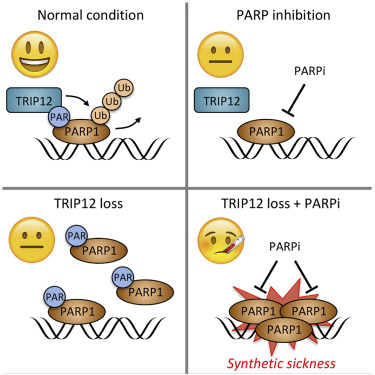
Gatti M, Imhof R, Huang Q, Baudis M, Altmeyer M
* equal contribution
The Ubiquitin Ligase TRIP12 Limits PARP1 Trapping and Constrains PARP Inhibitor Efficiency
Cell Reports 2020 Aug 4;32(5):107985.
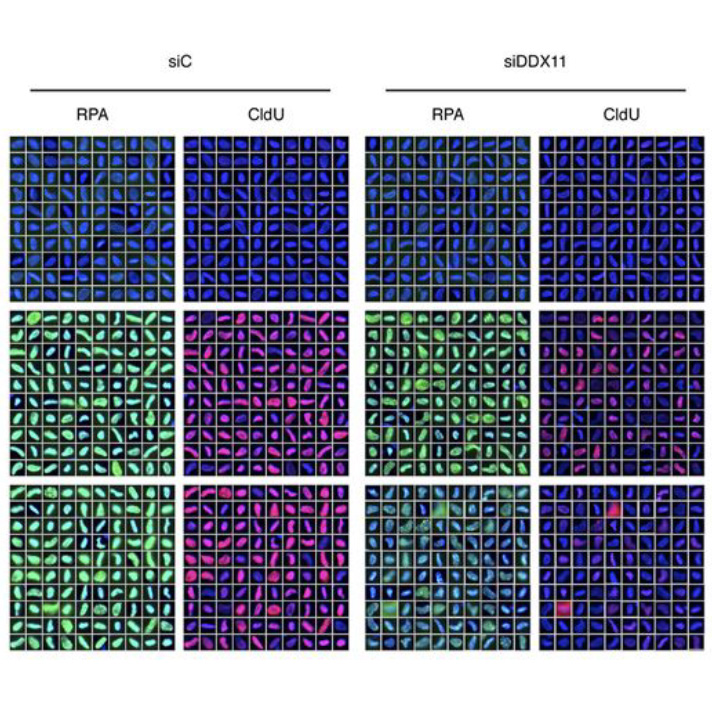
Simon A.K, Kummer S, Wild S, Lezaja A, Teloni F, Jozwiakowski S.K, Altmeyer M, Gari K
* equal contribution
The iron–sulfur helicase DDX11 promotes the generation of single-stranded DNA for CHK1 activation
Life Science Alliance, 2020 Feb 18;3(3):e201900547.
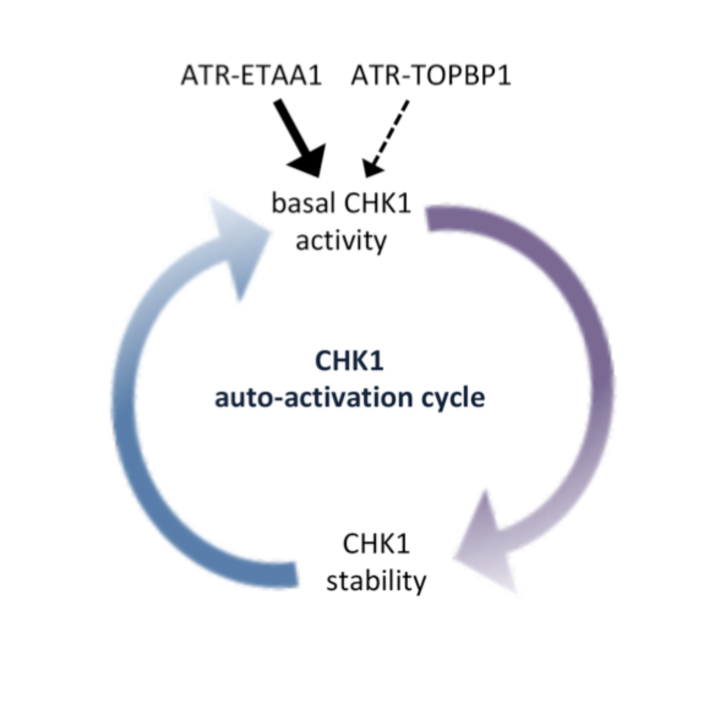
Michelena J, Gatti M, Teloni F, Imhof R, Altmeyer M
Basal CHK1 activity safeguards its stability to maintain intrinsic S-phase checkpoint functions
J Cell Biol. 2019 Jul 31. pii: jcb.201902085
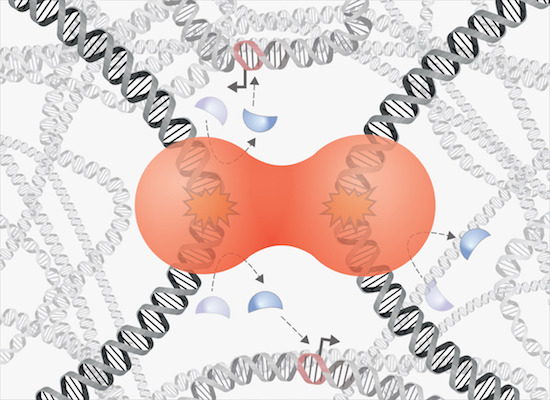
Kilic S, Lezaja A, Gatti M, Bianco E, Michelena J, Imhof R, Altmeyer M
Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments
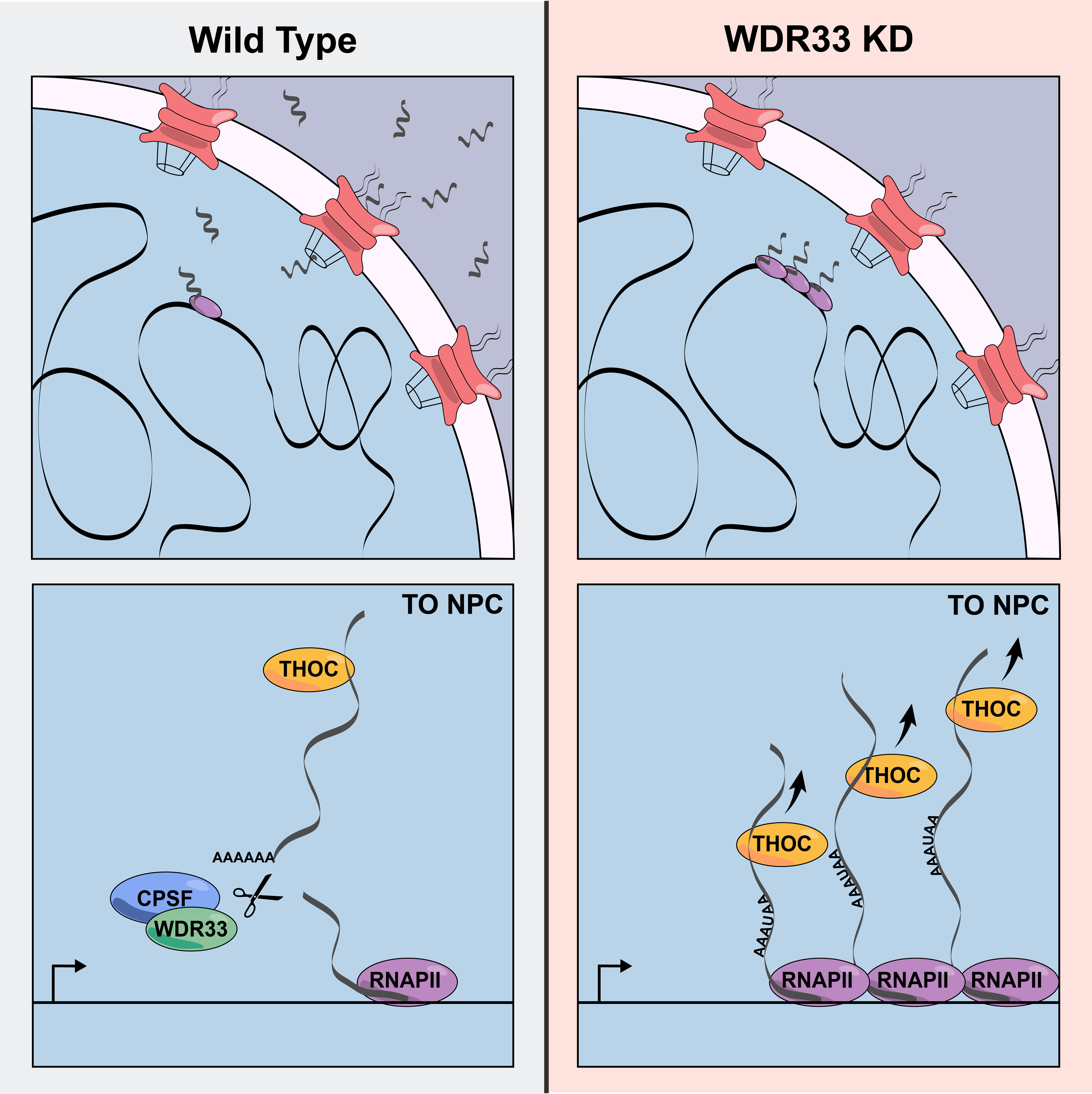
Teloni F, Michelena J, Lezaja A, Kilic S, Ambrosi C, Menon S, Dobrovolna J, Imhof R, Janscak P, Baubec T, Altmeyer M
Efficient Pre-mRNA Cleavage Prevents Replication-Stress-Associated Genome Instability
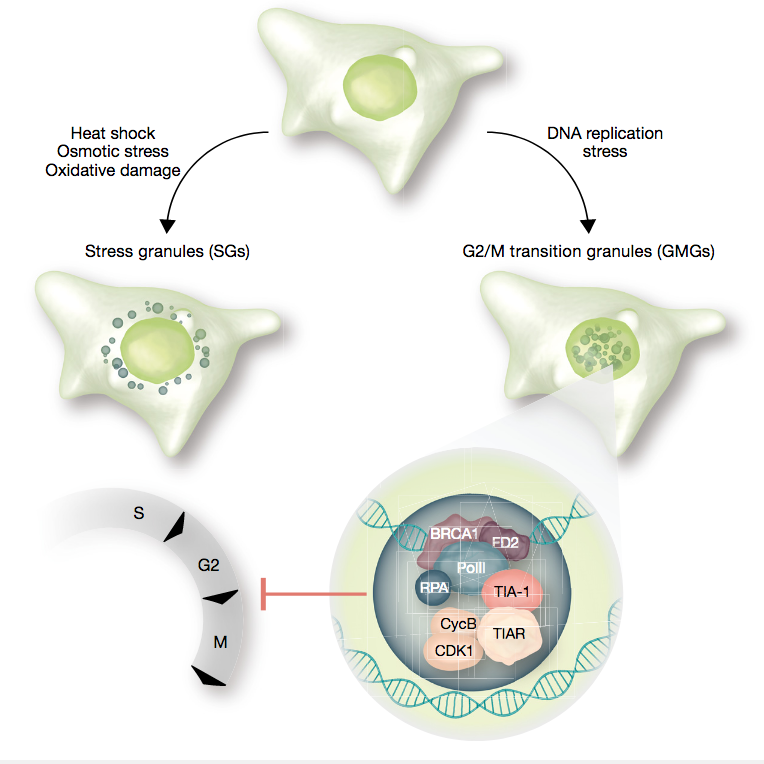
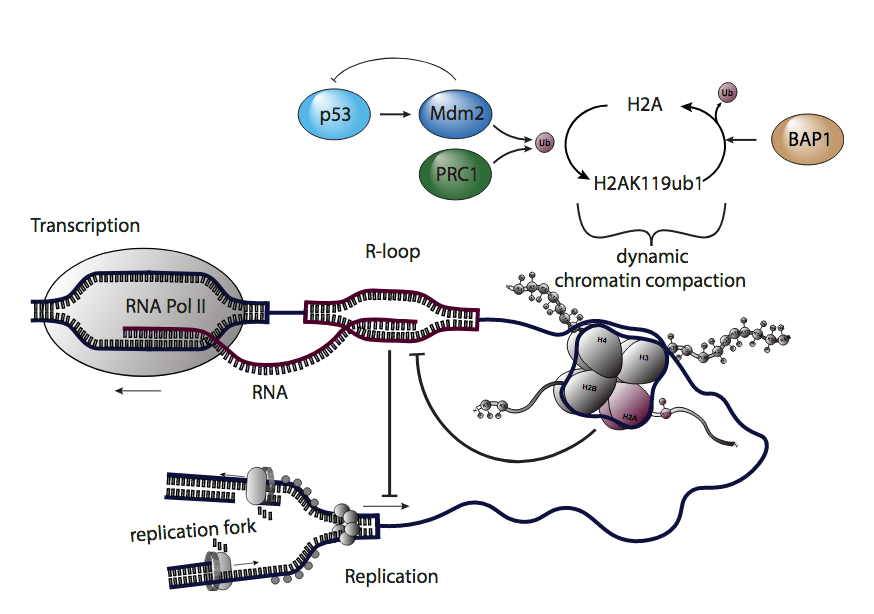
Klusmann I, Wohlberedt K, Magerhans A, Teloni F, Korbel JO, Altmeyer M, Dobbelstein M
Chromatin modifiers Mdm2 and RNF2 prevent RNA:DNA hybrids that impair DNA replication

Przetocka S, Porro A, Bolck HA, Walker C, Lezaja A, Trenner A, von Aesch C, Himmels SF, D'Andrea AD, Ceccaldi R, Altmeyer M, Sartori AA
CtIP-Mediated Fork Protection Synergizes with BRCA1 to Suppress Genomic Instability upon DNA Replication Stress
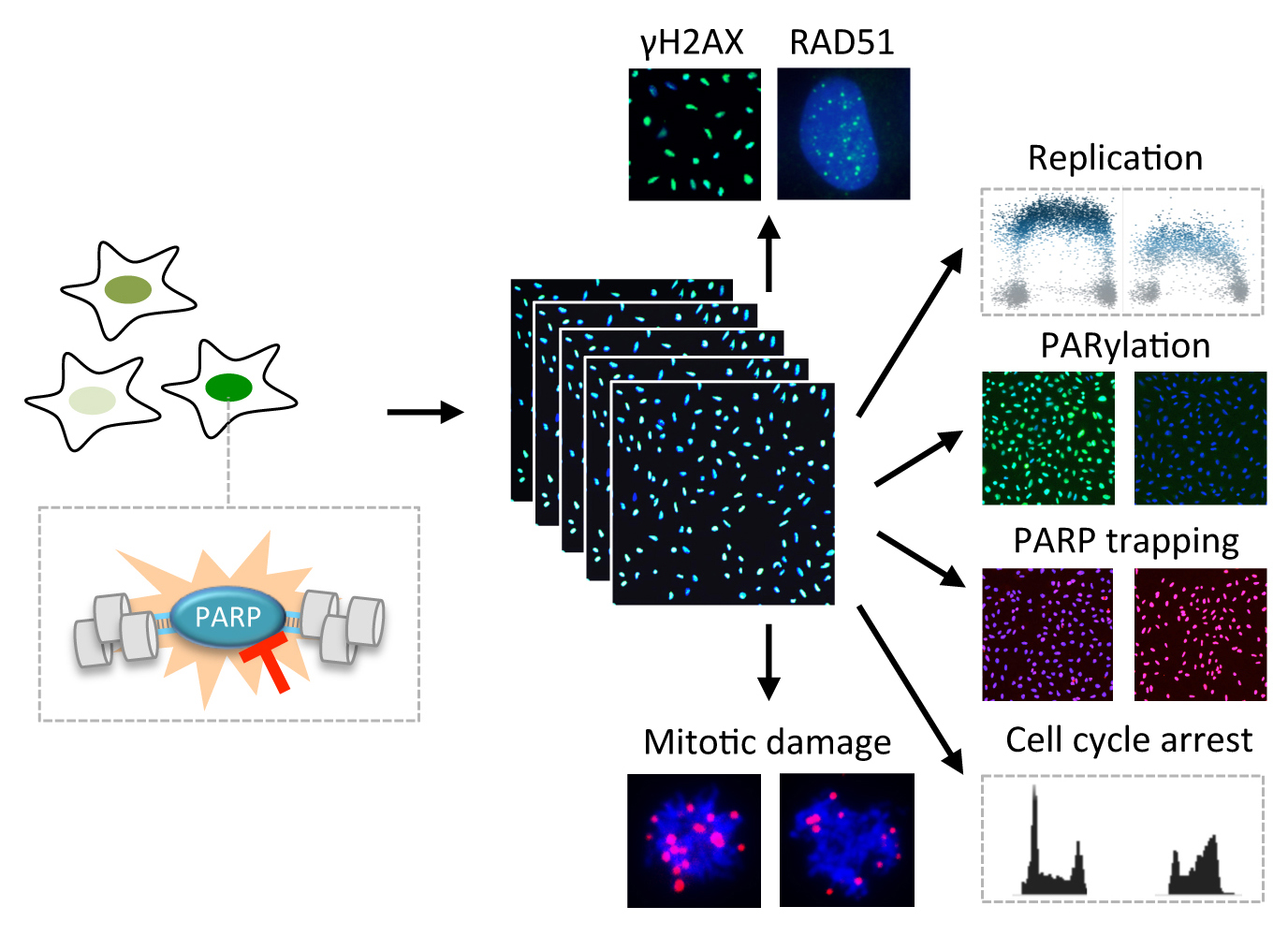
Michelena J, Lezaja A, Teloni F, Schmid T, Imhof R, Altmeyer M
Analysis of PARP inhibitor toxicity by multidimensional fluorescence microscopy reveals mechanisms of sensitivity and resistance
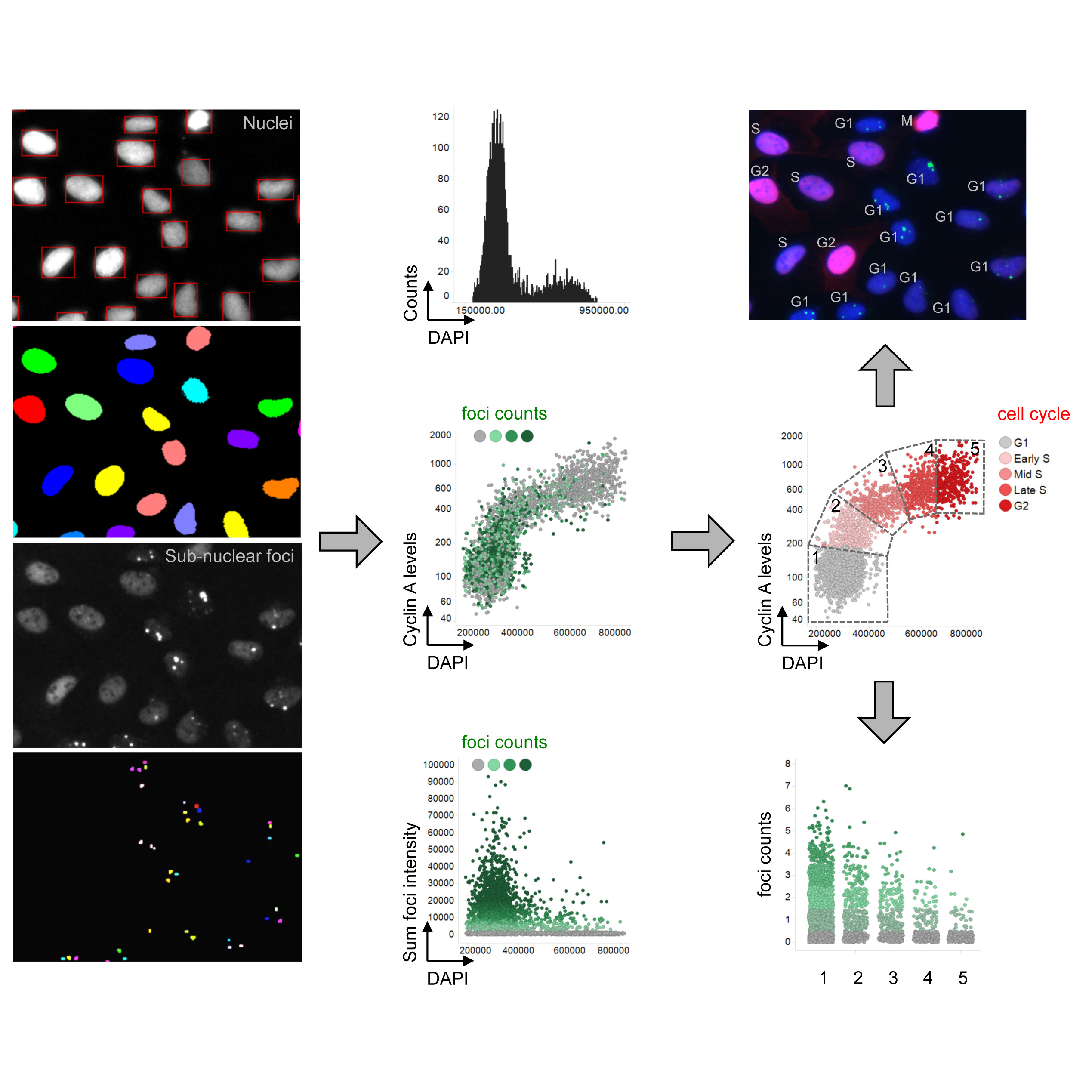
Lezaja A, Altmeyer M
High Content Imaging für Multidimensionale Zellanalysen
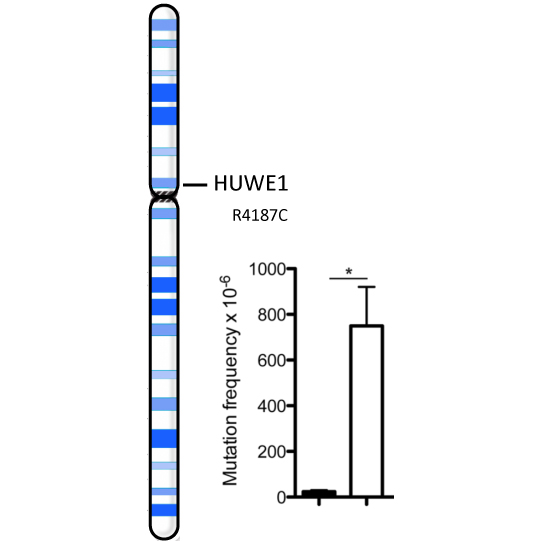
Bosshard M, Aprigliano R, Gattiker C, Palibrk V, Markkanen E, Hoff Backe P, Pellegrino S, Raymond FL, Froyen G, Altmeyer M, Bjørås M, Dianov GL, van Loon B
Impaired oxidative stress response characterizes HUWE1-promoted X-linked intellectual disability
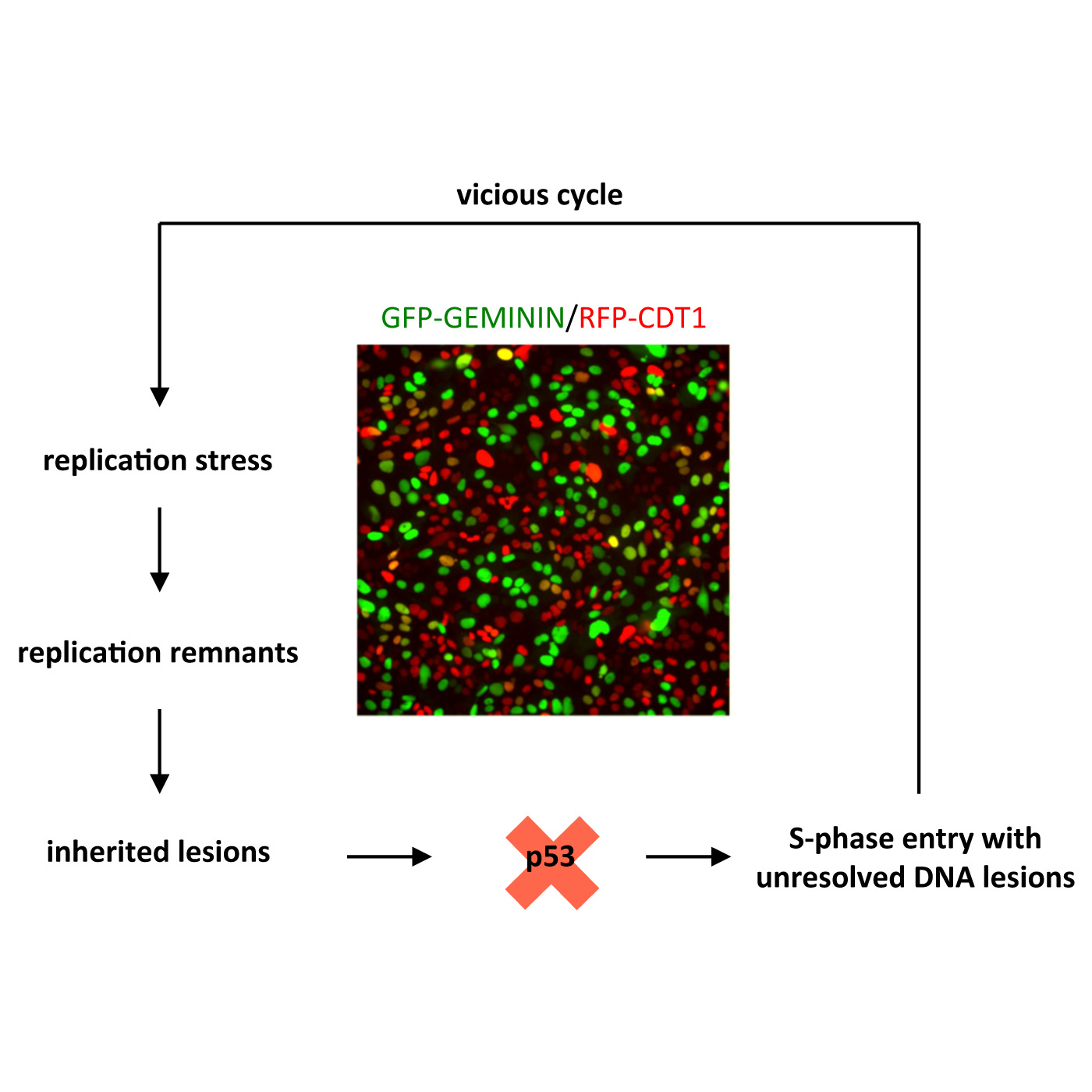
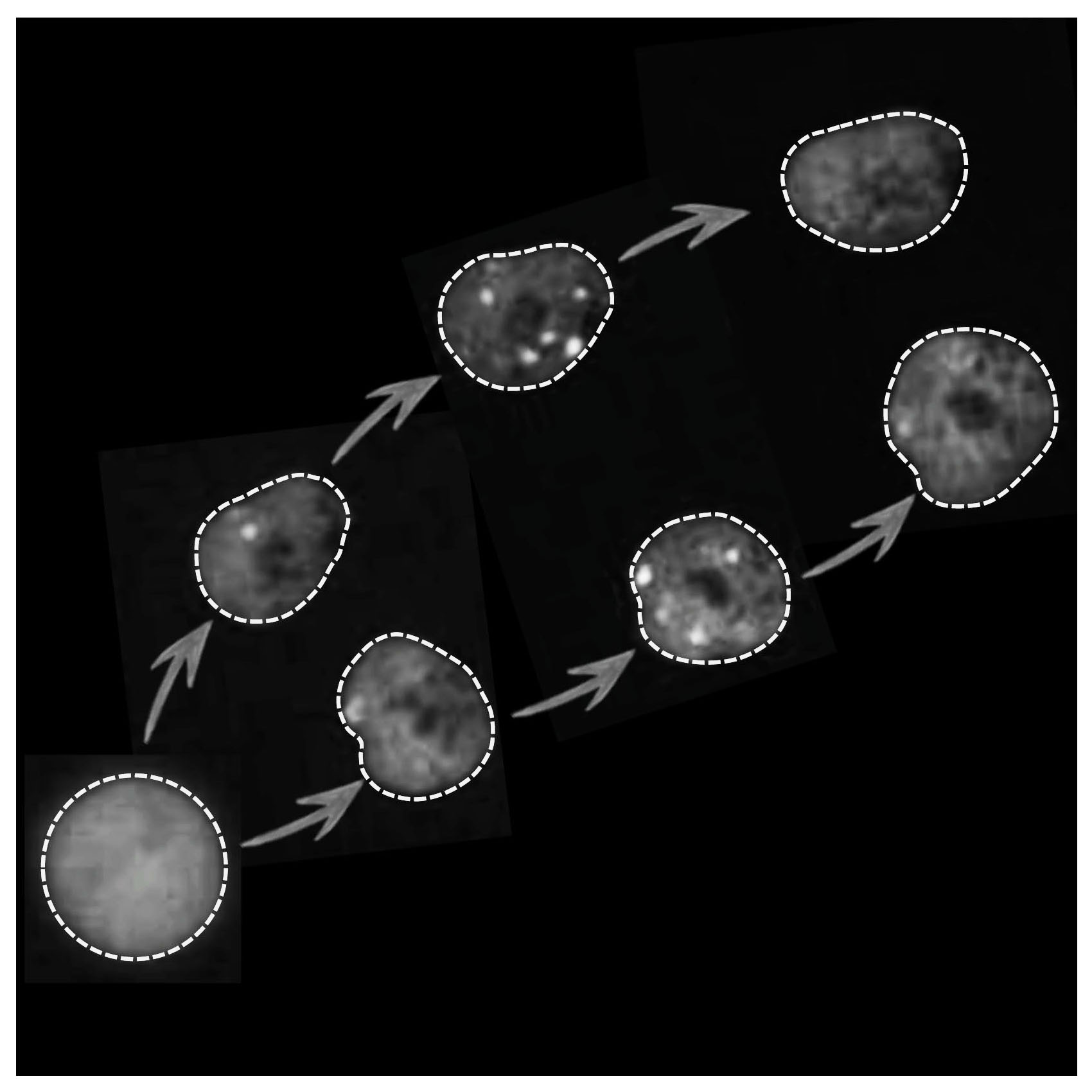
Lezaja A, Altmeyer M
Inherited DNA lesions determine G1 duration in the next cell cycle
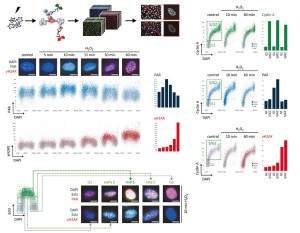
Michelena J, Altmeyer M
Cell cycle resolved measurements of poly(ADP-ribose) formation and DNA damage signaling by quantitative image-based cytometry
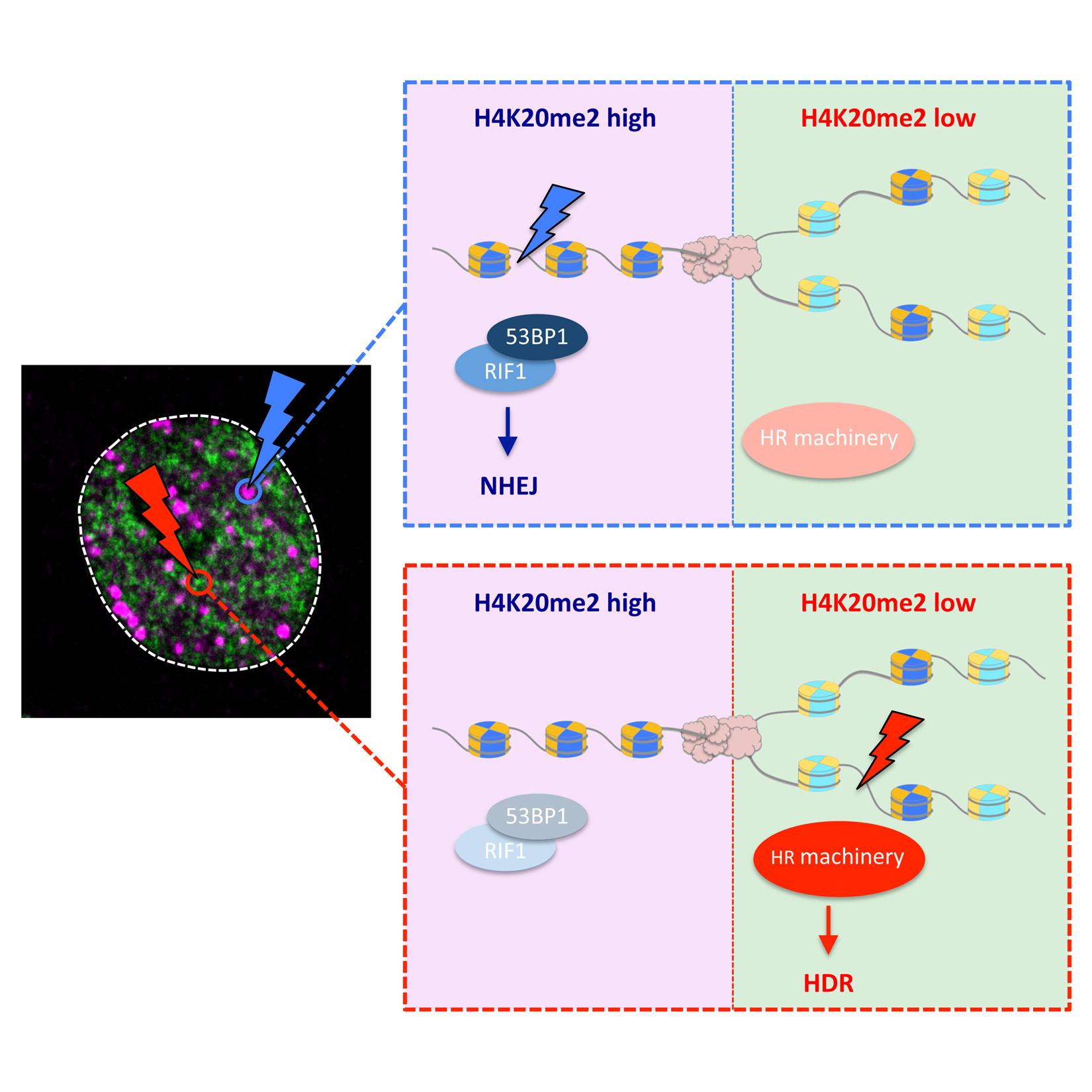
Pellegrino S* , Michelena J* , Teloni F, Imhof R, Altmeyer M
* equal contribution
Replication-Coupled Dilution of H4K20me2 Guides 53BP1 to Pre-replicative Chromatin
Cell Rep. 2017 May 30;19(9):1819-1831
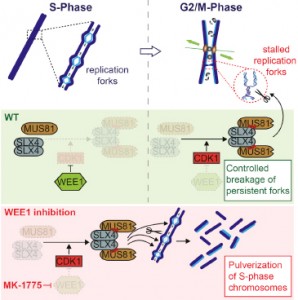
Duda H, Arter M, Gloggnitzer J, Teloni F, Wild P, Blanco MG, Altmeyer M, Matos J
A Mechanism for Controlled Breakage of Under-replicated Chromosomes during Mitosis
Developmental Cell 39, 740-755 December 19, 2016
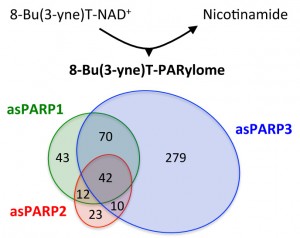
Mangerich A, Altmeyer M
Identifying ADP-ribosylation targets by chemical genetics
Translational Cancer Research. 2016
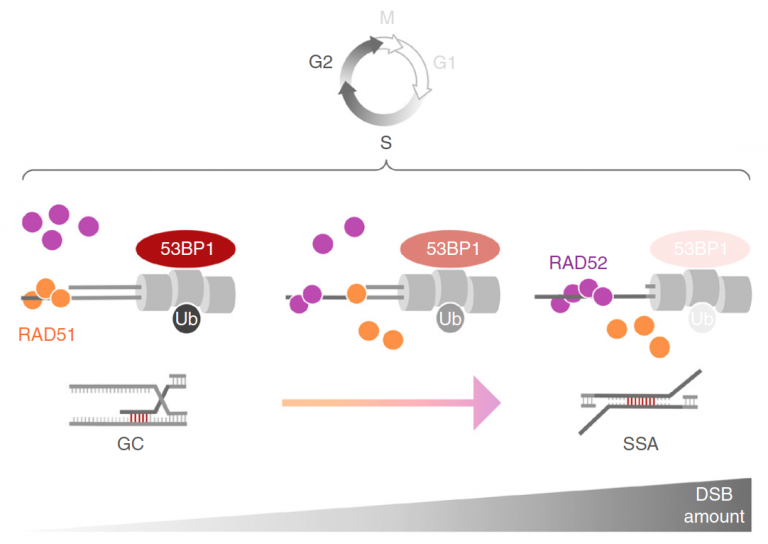
Ochs F, Somyajit K, Altmeyer M, Rask MB, Lukas J, Lukas C
53BP1 fosters fidelity of homology-directed DNA repair
Nat Struct Mol Biol. 2016 Aug;23(8):714-21
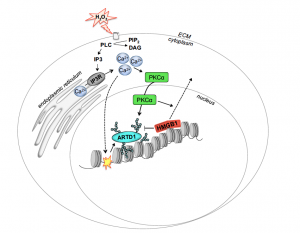
Andersson A, Bluwstein A, Kumar N, Teloni F, Traenkle J, Baudis M, Altmeyer M, Hottiger MO
PKCαand HMGB1 antagonistically control hydrogen peroxide-induced poly-ADP-ribose formation
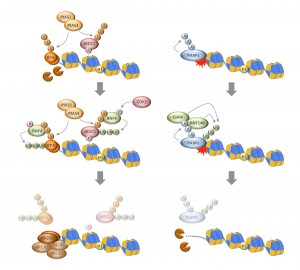
Pellegrino S, Altmeyer M
Interplay between Ubiquitin, SUMO, and Poly(ADP-Ribose) in the Cellular Response to Genotoxic Stress
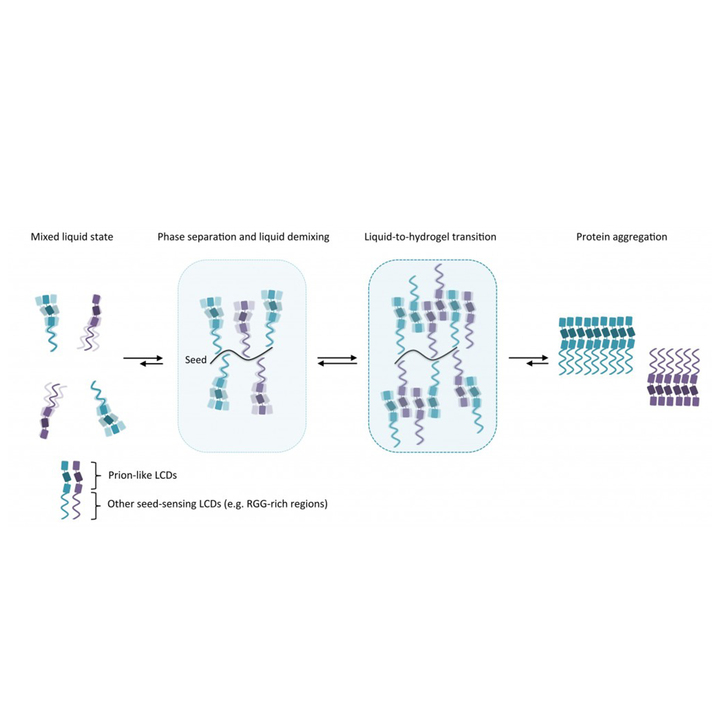
Aguzzi A, Altmeyer M
Phase Separation: Linking Cellular Compartmentalization to Disease
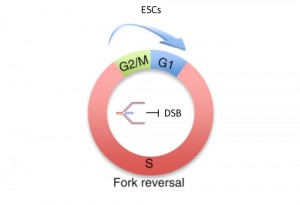
Ahuja AK, Jodkowska K, Teloni F, Bizard AH, Zellweger R, Herrador R, Ortega S, Hickson ID, Altmeyer M, Mendez J, Lopes M
A short G1 phase imposes constitutive replication stress and fork remodelling in mouse embryonic stem cells
Nat Commun. 2016 Feb 15;7:10660
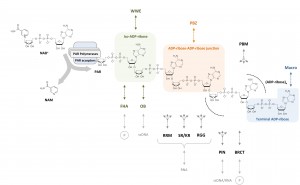
Teloni F, Altmeyer M
Readers of poly(ADP-ribose): designed to be fit for purpose
Nucleic Acids Res. 2016 Feb 18;44(3):993-1006
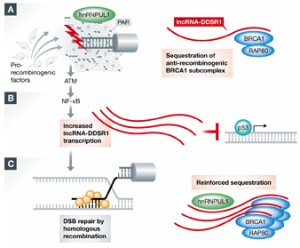
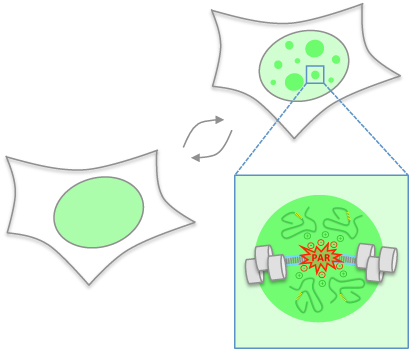
Altmeyer M" , Neelsen KJ, Teloni F, Pozdnyakova I, Pellegrino S, Grøfte M, Rask MB, Streicher W, Jungmichel S, Nielsen ML, Lukas J"
" co-corresponding
Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose)
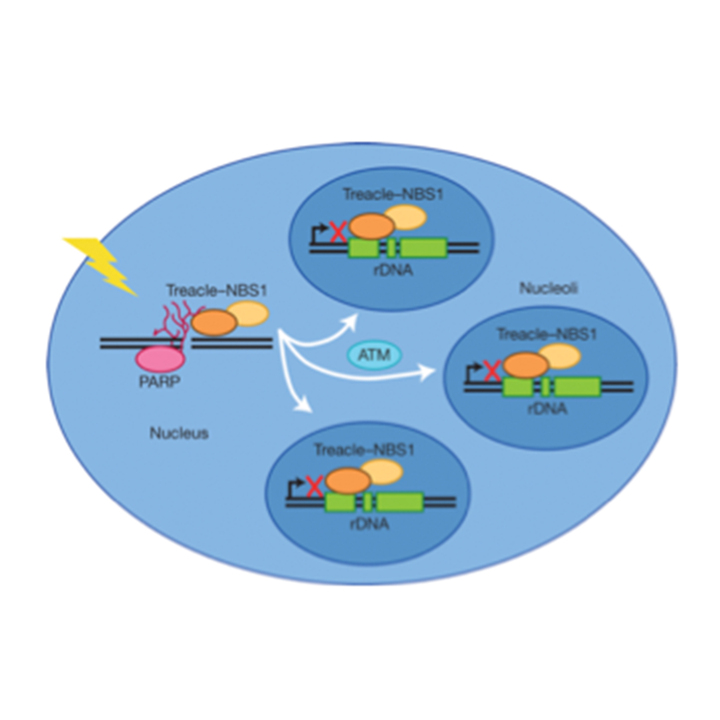
Larsen DH, Hari F, Clapperton JA, Gwerder M, Gutsche K, Altmeyer M, Jungmichel S, Toledo LI, Fink D, Rask MB, Grøfte M, Lukas C, Nielsen ML, Smerdon SJ, Lukas J, Stucki M
The NBS1-Treacle complex controls ribosomal RNA transcription in response to DNA damage
Nat Cell Biol. 2014 Aug;16(8):792-803

Engelken J, Altmeyer M, Franklin R
The disruption of trace element homeostasis due to aneuploidy as a unifying theme in the etiology of cancer
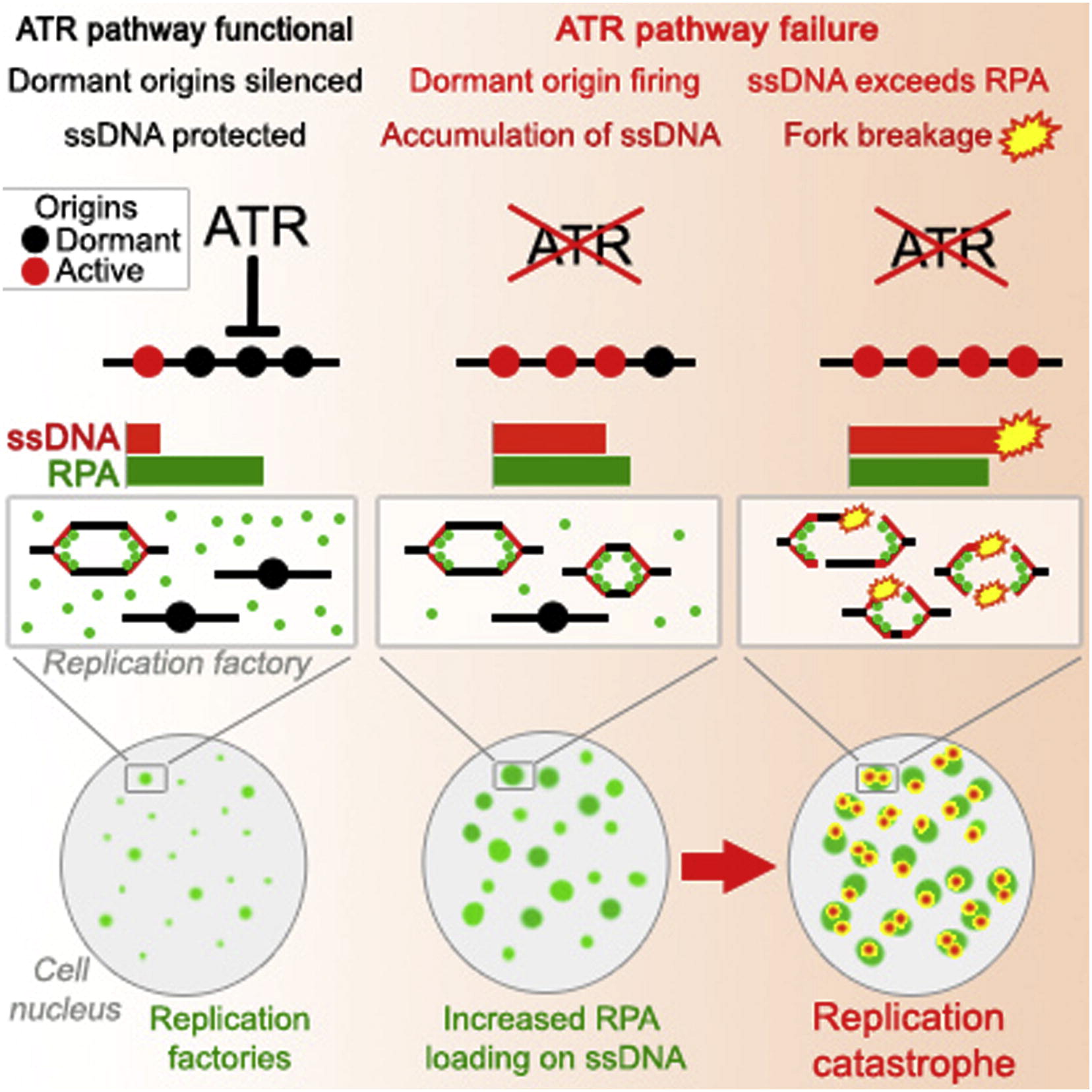
Toledo LI, Altmeyer M, Rask MB, Lukas C, Larsen DH, Povlsen LK, Bekker-Jensen S, Mailand N, Bartek J, Lukas J
ATR prohibits replication catastrophe by preventing global exhaustion of RPA

Jungmichel S, Rosenthal F, Altmeyer M, Lukas J, Hottiger MO, Nielsen ML
Proteome-wide identification of poly(ADP-Ribosyl)ation targets in different genotoxic stress responses
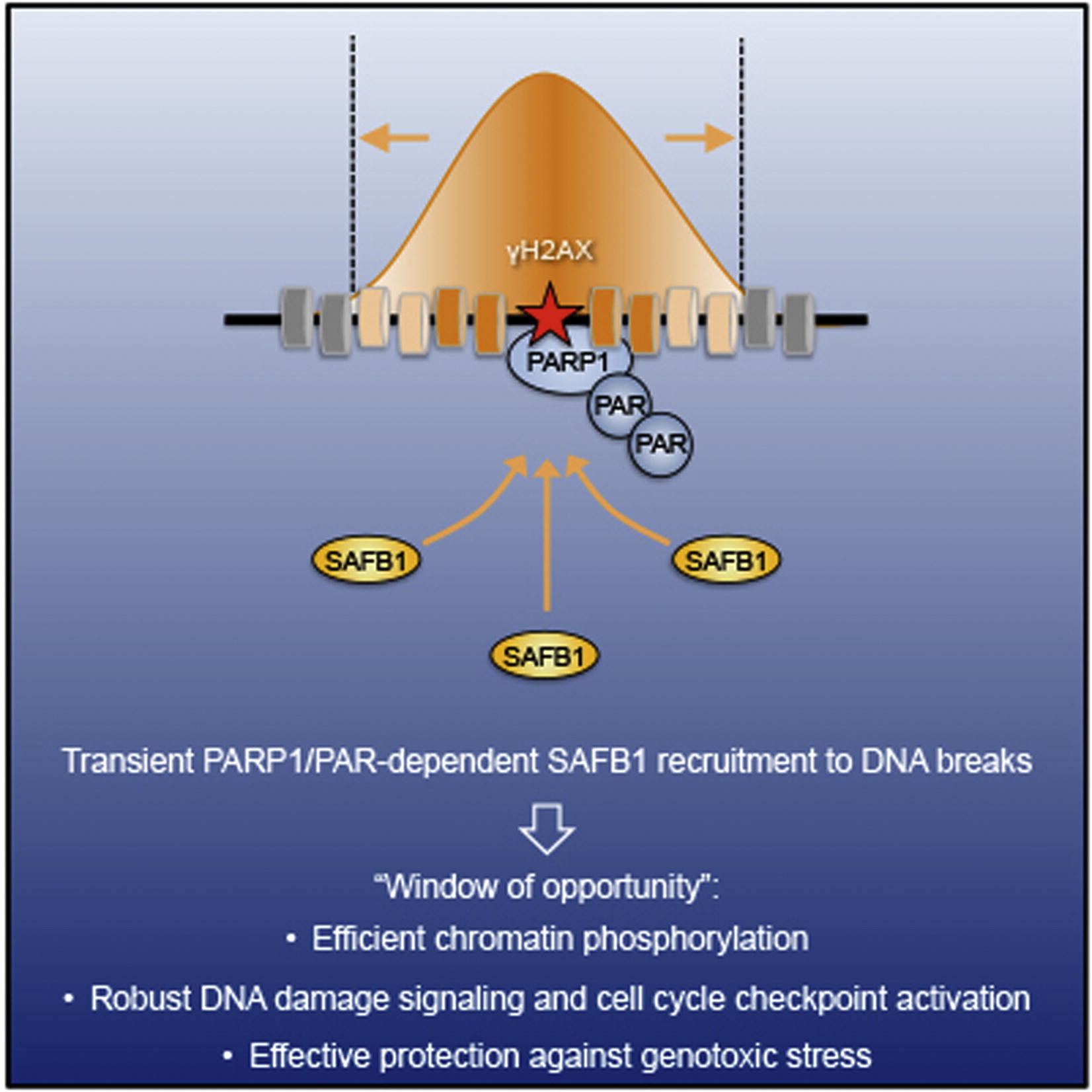
Altmeyer M, Toledo L, Gudjonsson T, Grøfte M, Rask MB, Lukas C, Akimov V, Blagoev B, Bartek J, Lukas J
The chromatin scaffold protein SAFB1 renders chromatin permissive for DNA damage signaling
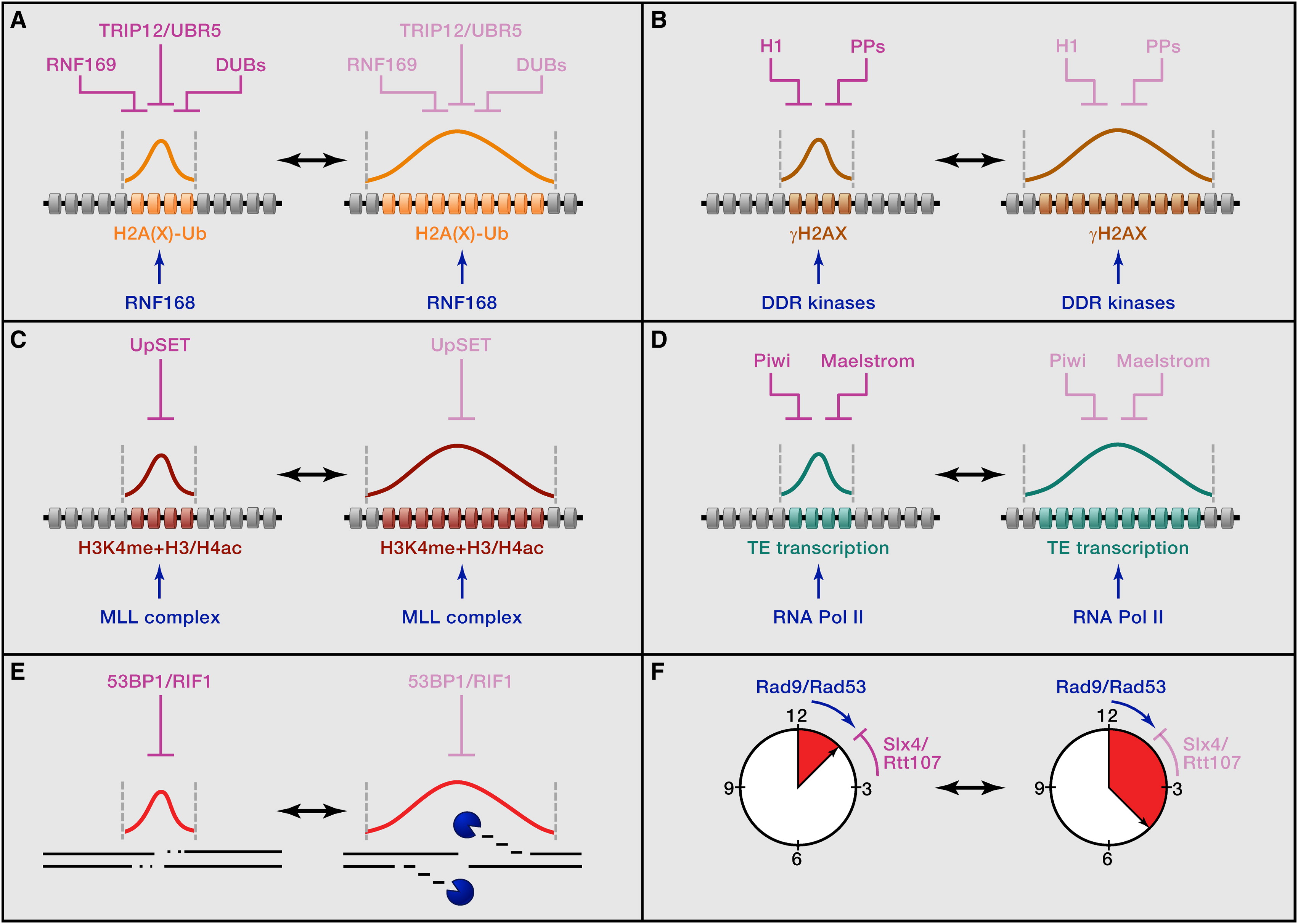
Altmeyer M", Lukas J"
" co-corresponding
Guarding against collateral damage during chromatin transactions
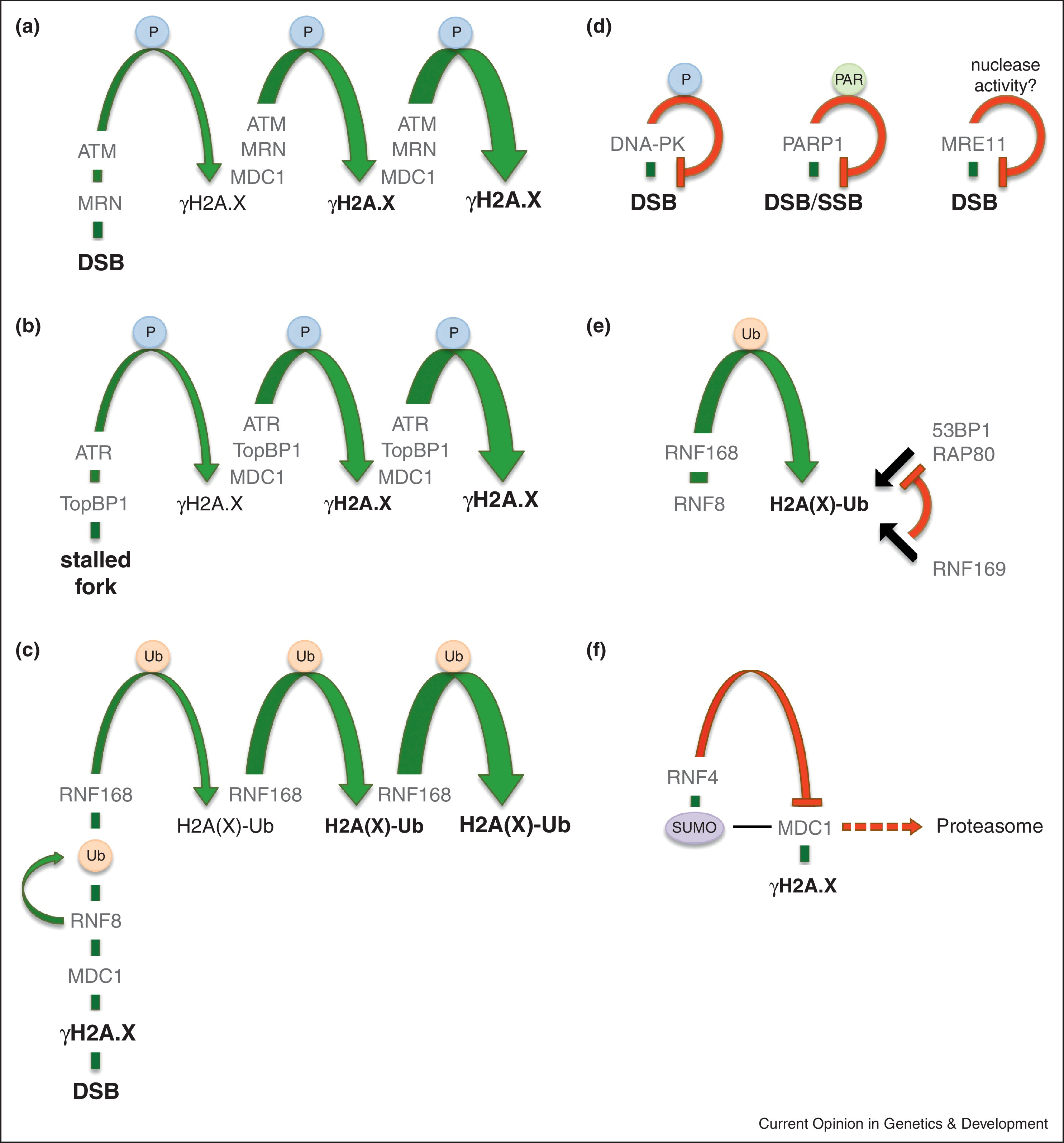
Altmeyer M", Lukas J"
" co-corresponding
To spread or not to spread--chromatin modifications in response to DNA damage
Curr Opin Genet Dev 2013;23(2):156-65
Altmeyer M
Addicted to PAR? A closer look at PARP inhibitor sensitivity
Cell Cycle. 2012 Nov 1;11(21):3916
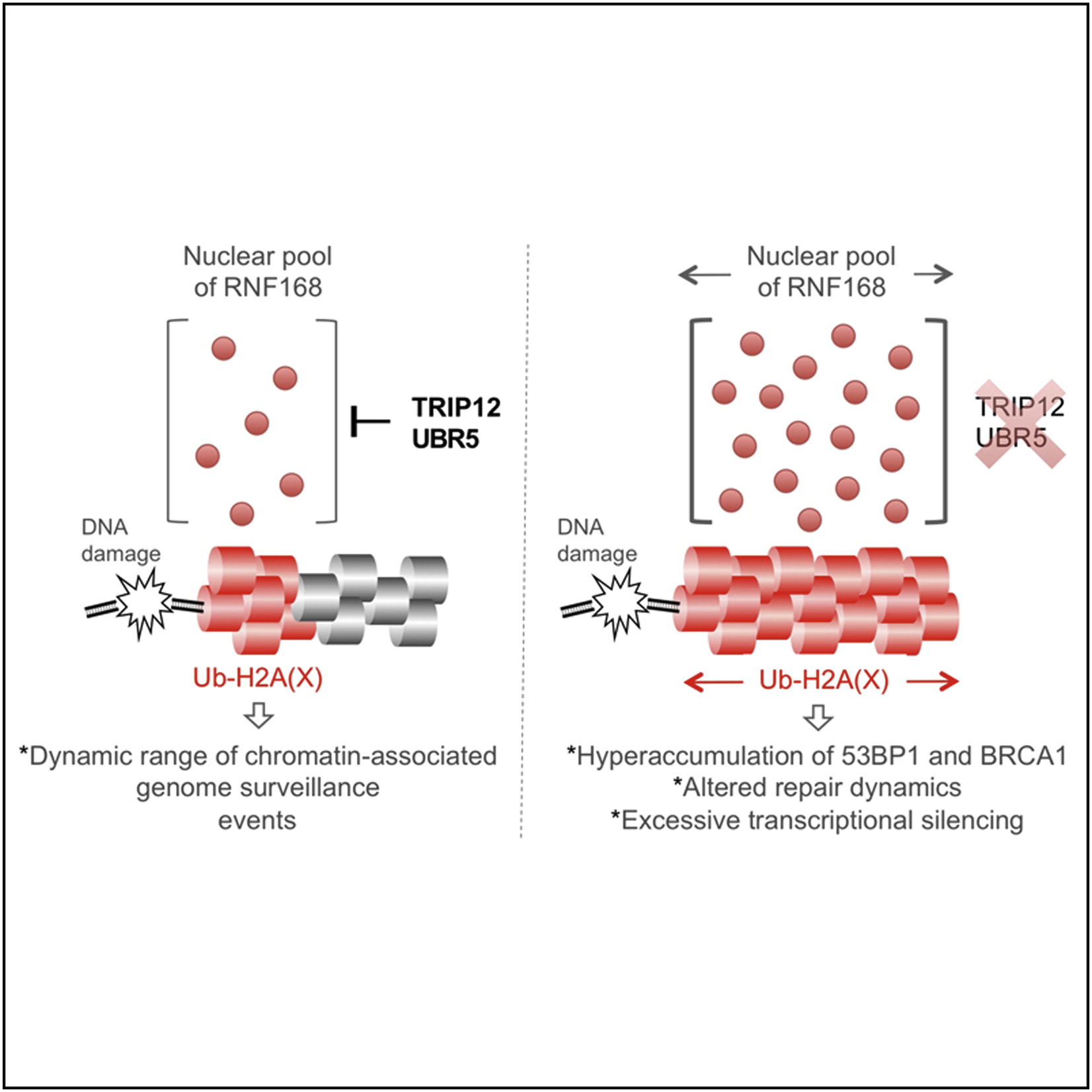
Gudjonsson T*, Altmeyer M*, Savic V, Toledo L, Dinant C, Grøfte M, Bartkova J, Poulsen M, Oka Y, Bekker-Jensen S, Mailand N, Neumann B, Heriche JK, Shearer R, Saunders D, Bartek J, Lukas J, Lukas C
* equal contribution
TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes
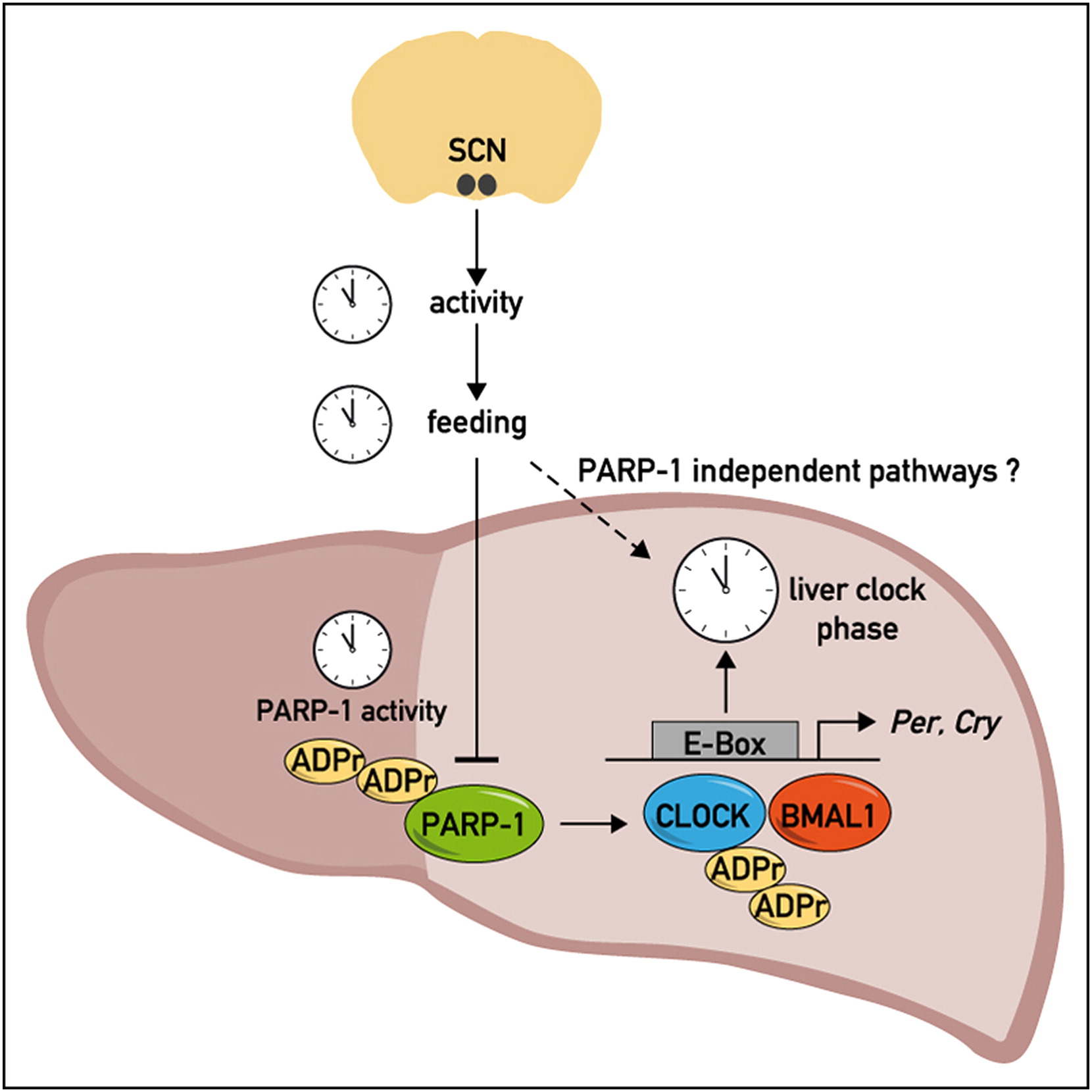
Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, Hottiger MO, Schibler U
Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding
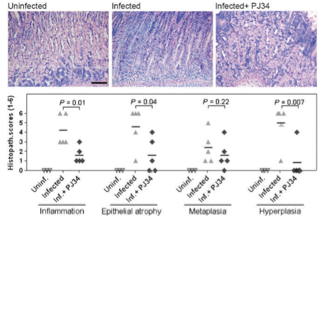
Toller IM, Altmeyer M, Kohler E, Hottiger MO, Müller A
Inhibition of ADP ribosylation prevents and cures helicobacter-induced gastric preneoplasia
Cancer Res. 2010 Jul 15;70(14):5912-22
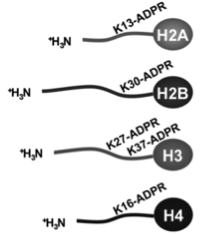
Messner S*, Altmeyer M*, Zhao H, Pozivil A, Roschitzki B, Gehrig P, Rutishauser D, Huang D, Caflisch A, Hottiger MO
* equal contribution
PARP1 ADP-ribosylates lysine residues of the core histone tails
Nucleic Acids Res. 2010 Oct;38(19):6350-62.
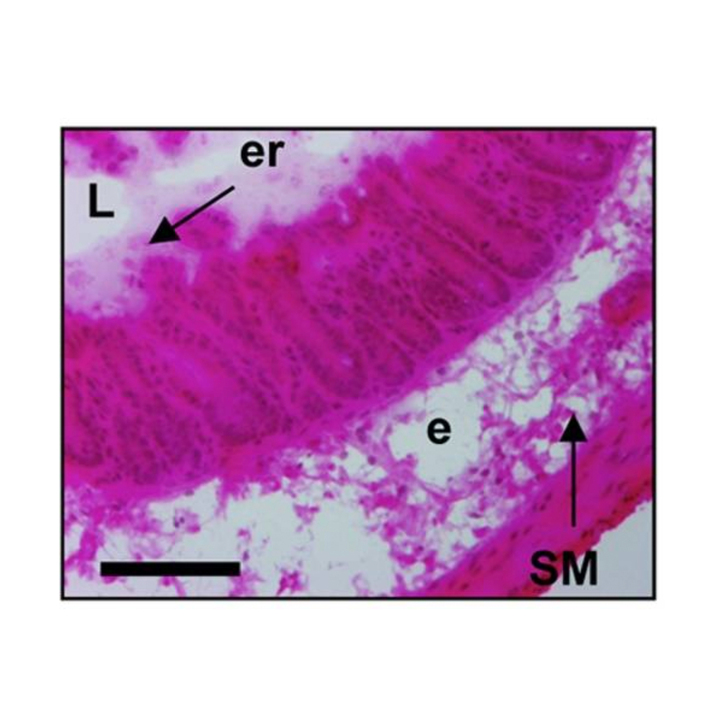
Altmeyer M, Barthel M, Eberhard M, Rehrauer H, Hardt WD, Hottiger MO
Absence of poly(ADP-ribose) polymerase 1 delays the onset of Salmonella enterica serovar Typhimurium-induced gut inflammation
Infect Immun. 2010 Aug;78(8):3420-31
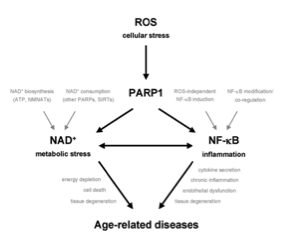
Altmeyer M, Hottiger MO
Poly(ADP-ribose) polymerase 1 at the crossroad of metabolic stress and inflammation in aging
Aging (Albany NY). 2009 May 20;1(5):458-69
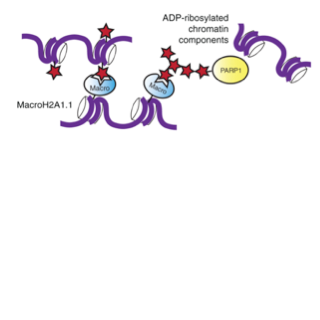
Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EH, Scheffzek K, Hottiger MO, Ladurner AG
A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation
Nat Struct Mol Biol. 2009 Sep;16(9):923-9
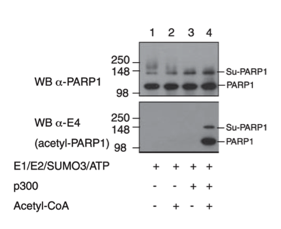
Messner S, Schuermann D, Altmeyer M, Kassner I, Schmidt D, Schär P, Müller S, Hottiger MO
Sumoylation of poly(ADP-ribose) polymerase 1 inhibits its acetylation and restrains transcriptional coactivator function
FASEB J. 2009 Nov;23(11):3978-89
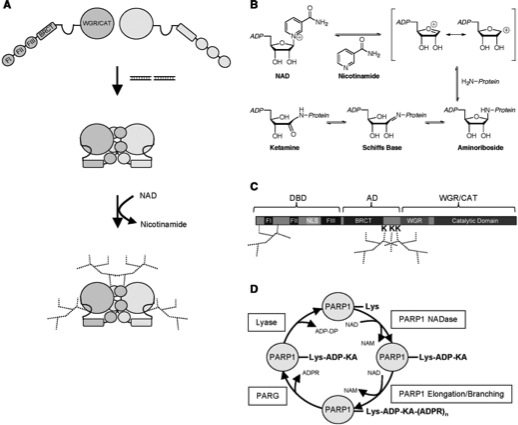
Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO
Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites
Nucleic Acids Res 2009;37(11):3723-38
Haenni SS*, Altmeyer M*, Hassa PO*, Valovka T, Fey M, Hottiger MO
* equal contribution
Importin alpha binding and nuclear localization of PARP-2 is dependent on lysine 36, which is located within a predicted classical NLS
BMC Cell Biol. 2008 Jul 21;9:39.
Haenni SS*, Hassa PO*, Altmeyer M*, Fey M, Imhof R, Hottiger MO
* equal contribution
Identification of lysines 36 and 37 of PARP-2 as targets for acetylation and auto-ADP-ribosylation
Int J Biochem Cell Biol. 2008;40(10):2274-83.

Fahrer J, Kranaster R, Altmeyer M, Marx A, Bürkle A
Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length
Nucleic Acids Res. 2007;35(21):e143